FARSIGHT Framework
| Line 1: | Line 1: | ||
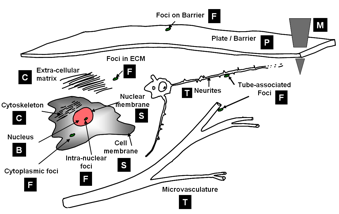
| − | We start by assuming that we are given one or more multi-dimensional image(s) <math>I(x,y,z,\lambda,t)</math>, as appropriate. The hardest first step to automated image analysis is image segmentation. We simplify this task | + | We start by assuming that we are given one or more multi-dimensional image(s) <math>I(x,y,z,\lambda,t)</math>, as appropriate. The hardest first step to automated image analysis is image segmentation. We simplify this task using a 'divide and conquer' strategy. We recognize that some of the best-available segmentation algorithms We are primarily interested in fluorescence microscopy at the cell and tissue level <math>(0.1 - 1000 \mu m)</math>. Within this realm, we have found that, in spite of the variability in biological forms, it is possible to identify a “short list” of frequently occurring morphologies of biological entities – blobs (B), tubes (T), shells (S), foci (F), plates (P), clouds (C), and man-made objects (M). These morphologies are illustrated in the following figure: |
| + | [[Image:CTM.png]] | ||
Revision as of 19:57, 9 April 2009
We start by assuming that we are given one or more multi-dimensional image(s) I(x,y,z,λ,t), as appropriate. The hardest first step to automated image analysis is image segmentation. We simplify this task using a 'divide and conquer' strategy. We recognize that some of the best-available segmentation algorithms We are primarily interested in fluorescence microscopy at the cell and tissue level (0.1 − 1000μm). Within this realm, we have found that, in spite of the variability in biological forms, it is possible to identify a “short list” of frequently occurring morphologies of biological entities – blobs (B), tubes (T), shells (S), foci (F), plates (P), clouds (C), and man-made objects (M). These morphologies are illustrated in the following figure: