Glossary
Some of the words used to describe FARSIGHT are defined here:
Contents |
Image
An image is understood to be a point-by-point spatial map of one or more physical properties of the specimen under the microscope. For our purposes, an image is a rectangular array of values. For 2-D images, each point is termed a pixel. For 3-D images, each point is termed a voxel. Each pixel corresponds to a small spatial region of the specimen with physical dimensions (Δx,Δy). Each voxel is associated with dimensions (Δx,Δy,Δz). Most microscopy derived images are non-isotropic, meaning that two or more of the physical dimensions are unequal. Usually, the lateral dimensions are equal, i.e., Δx = Δy, but the axial dimension is much greater, i.e., Δz > Δy. In addition to physical dimensions, each pixel/voxel is also associated with an intensity value (a.k.a. grayscale value) that is stored as a N − bit number (a bit is a binary digit). An intensity value that is stored as a N − bit value can be as low as 0 and as high as (2N − 1). Its dynamic range is 2N.
Image Metadata
This is a catch-all phrase used to describe information about an image. This data is stored as a XML file in the OME-TIFF file format, and incorporated into the image header. Image file headers can be quite simple or elaborate. They can store information on where the data came from (its provenance), microscope settings user to record the image, etc. The BioFormats package in FARSIGHT and the OMERO image database provide tools to view, and edit image metadata.
Image Intensity
This term refers to the brightness of a point in an image. The intensity of a pixel is determined by several quantities including the local concentration of a fluorophore, the quantum efficiency of the fluorophore, sensitivity of the light sensor in the imaging system, etc. Most fluorophores are merely labels that attach themselves to molecules of biological interest. In other words, fluorescence intensity is an indirect indicator of the biochemical that we are interested in imaging. This latter relationship is commonly non-proportional due to the use of fluorescence amplification techniques that are impossible to control precisely. Overall, it is important keep in mind is that the image intensity is rarely a stoichiometric indicator. In other words, one cannot infer the molar concentration of the fluorophore from the image intensity.
Object Appearance
This is a term from the field of image analysis that refers to the pattern of image brightness (intensity) variations across an object. The texture of an object is a good example of such a property
Time-lapse Movie
A time-lapse movie is a sequence of snapshots of a live specimen that is undergoing changes. The sampling interval is the physical time Δt between successive snapshots. Each image in a time-lapse series could be a multi-dimensional one.
Hyper-spectral & Multi-spectral Images
In a hyper-spectral image, one records an entire finely-resolved absorption/emission spectrum of the specimen at each pixel s(x,y,λ) with a uniform spectral sampling unit Δλ. When the spectrum is sampled non-uniformly, the term multi-spectral cube is often employed to describe such data. A range of wavelengths is referred to as a band.
Multi-Channel Images
An image made up of multiple channels is called a Multi-Channel Image. A very common example of a Multi-Channel Image is an RGB image which has three channels: red, green, and blue.
Imaging
This is the process by which an image is acquired (recorded). Imaging is the only way to record spatial location, structure, and context information, basically anything of a “spatial nature”.
Molecular Imaging
This is a term used for imaging specific biochemicals with high specificity. This type of imaging is usually performed using a fluorescent tag or label. The fluorescent tag attaches itself with high specificity to the substance of interest, and reveals itself by emitting fluorescent light when excited optically. Molecular imaging is not to be confused with the idea of single-molecule imaging.
Multi-modality Imaging
The word "modality" refers to an imaging method that measures one particular physical property at each pixel/voxel. Some microscopes can measure more than one property (e.g., phase & fluorescence). Such imaging systems are termed multi-modal. each pixel/voxel of a multi-modal image contains a vector of measurements, one for each modality. each element of this vector for the whole image is termed a channel.
High-throughput Imaging
The word throughput means the number of images that can be captured per hour. High-throughput microscopes are optimized for throughput. The related term "high-throughput experimentation" implies that a large number of experiments are achieved per hour. In the world of imaging, this translates to the idea of parallel experimentation. For instance, instead of studying one specimen at a time, one can study an entire array of specimens in a micro-well plate. These plates can hold hundreds of specimens in tiny wells that are precisely spaced. The high-throughput (HT) microscope is equipped with automated hardware for systematically moving from one micro-well to the next, and recording images from each. These systems must also possess other types of hardware, for example autofocus systems to eliminate the need for manual intervention. One can vary the experimental parameters in a systematic manner across the array. Many contemporary high-throughput imaging systems also permit high-content imaging.
High-content Imaging
This is the method of trying to extract much more information from specimens. Usually, this is accomplished by using more than one fluorescent channel to acquire information on multiple molecular markers of interest. As noted above, this type of imaging is also conducted in a high-throughput manner. The acronym HTHC stands for high-throughput high-content imaging. HTHC systems require considerable computational hardware and software to analyze the large amounts of data that are collected rapidly.
Channel
The image data from each fluorescent label or imaging modality is commonly referred to as a “channel”. Think of it as a monochrome image that is part of a more complete multi channel image. One type of channel of particular interest to the FARSIGHT project is a pure channel that contains only one type of biological object. This happens when the fluorescent label is sufficiently specific to the object, and there is negligible spectral overlap (“cross talk”) with other channels. Pure channels are valuable because they are particularly amenable to automated analysis. It is possible to develop specialized segmentation algorithms for each type of “object”. These are dramatically simpler than multi-channel segmentation since they only need to be able to handle one object type at a time. High-performance segmentation is possible in this case since we can exploit the specialization to develop highly reliable segmentation algorithms.
Impure Channel
A channel that is not pure. Basically, there are two kinds of impurities to consider: (i) Morphologically impure - this is the case when we have 2 or more types of objects in a single channel; and (ii) Spectrally impure - this is the case when the spectra of two or more fluorescent labels overlap and they cannot be untangled. Objects in impure channels are harder to segment, but not impossible. Many segmentation algorithms are model based, and only delineate objects that fit the model closely. Other objects can be separated out, and segmented by another algorithm. Spectrally pure channels can often be cleaned out using well-designed spectral unmixing algorithms, and adjustments to specimen preparation and imaging steps.
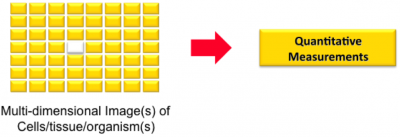
Image Analysis
This is the process (manual/automated) by which one extracts geometric descriptions and one or more quantitative measurements from an image. Think of it as "image(s) in, measurements out". Think of it also as the reverse of computer graphics (a.k.a. image synthesis) "geometric description in, image out".
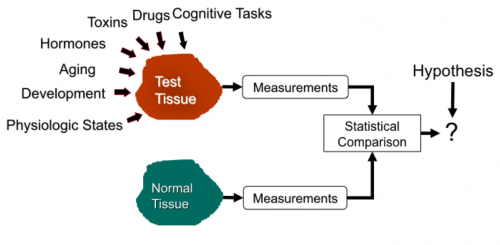
Image-based measurements are widely needed in the life sciences. Perhaps the most widespread need is to test biological hypotheses. The following diagram illustrates the use of image-based measurements for hypothesis testing. The biological researcher forms a scientific hypothesis about the result of a specific perturbation to a cell/tissue, keeping all other factors unchanged. Image-based measurements serve to confirm or defeat the hypothesis. The result is a unit of new knowledge that is combined with the existing body of knowledge. This act usually leads to more hypotheses to be tested. This is the method of science.
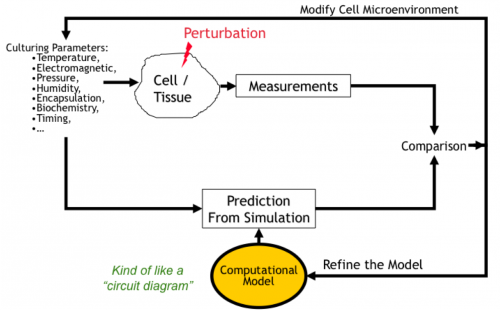
Increasingly, biologists are building predictive computational models of biological systems. This is the field of systems biology. Image-based measurements are needed to compare a computationally predicted behavior against actual observed behavior. Any observed discrepancies help scientists to refine their models, and this process is repeated for a variety of perturbations until a sufficiently descriptive model is attained.
Image Processing
This is the process by which an image is processed computationally to generate yet another image. Think of it as "image(s) in, image(s) out". Image smoothing is a simple example of image processing.
Unbiased Stereology
This is a set of manual image analysis methods that were invented before modern computational image analysis methods were developed. These methods continue to be used widely, and software tools are currently available to implement stereological image analysis protocols. These methods work by providing systematic ways to subsample the image data, and extrapolate from manual analysis of subsampled regions. As the name implies, these methods strive to minimize the statistical bias associated with the subsampling. In order to achieve this admirable and important goal, several assumptions are made. One such assumption is that the tissue is homogeneous (this is rarely the case in reality). Even when the achieved bias is low, it is possible for the variance to be high. Lowering the variance requires analysis of many more tissue samples. Indeed, some stereologists advocate extraction of small numbers of measurements across large numbers of specimens. Finally, stereological methods are poorly suited to multi-dimensional data and analysis of associations.
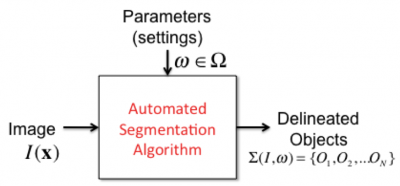
Segmentation
We use this catch-all word to refer to the procedure for delineating biological objects in an image. We are largely interested in automated and semi-automated segmentation methods (click here for some comments on manual segmentation methods). The purpose of such delineation is to enable image-based measurements of the biological objects (e.g., object sizes, shapes, and distances). The delineation can occur in several ways. The simplest is to assign a segmentation label to each point in the image (pixel / voxel). Groups of pixels with the same label constitute an object whose Object identifier (ID) is equal to the label value. More sophisticated forms of object delineation include geometric descriptions of the objects. For example, triangulated mesh representations of the object surfaces, and polyline representations of object centerlines (e.g., traces of vessel/neurites). Overall, segmentation takes us from an unstructured array of image points (pixels/voxels) to higher-level representations representing objects. It establishes a higher level of abstraction for further image analysis. Segmentation is perhaps the hardest step in automated image analysis, and a perfect segmentation algorithm is yet to be invented. It remains the subject of research. The goal of the FARSIGHT project is to incorporate best-available segmentation algorithms, and the means to simplify the task of segmentation to the extent possible, keeping in mind the needs of microscopy data.
Parameters
We use this word to refer to the user-specifiable settings for an algorithm. To get a correct and accurate output, it is usually necessary to choose the parameters optimally. Although it would be nice to have algorithms that do not require such parameters, the current reality is that most algorithms do require them. For instance, if you want to invoke an image smoothing algorithm, you need a way to also specify the degree of smoothing desired. Algorithms may be more or less sensitive to parameters. An algorithm is said to be sensitive if small adjustments to parameters cause significant changes to the outputs, and vice versa. Usually, parameters cannot have arbitrary values, and can only be adjusted over a defined range. Within this range, we often have preferred values that work "most of the time" - we call them default parameters. The goal of the FARSIGHT project is to develop ways to optimize parameters and simplify/automate this process to the extent possible.
Features
This is a term from the field of computer vision / image analysis that refers to quantitative measurements made from images. Usually, we are interested in features of biological objects (organelles, cells, neurites, vessels, etc.). Features are often the measurements of ultimate interest from image analysis. We also compute and use features that are only used to aid image analysis steps. One class of such features are termed diagnostic features in FARSIGHT. These features serve to detect/highlight segmentation errors, for example. In FARSIGHT, we are interested in two broad classes of end-user features: intrinsic features, and associative features.
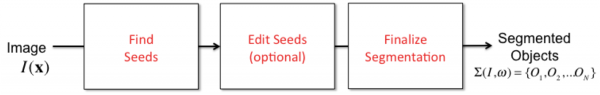
Seeds
Some segmentation algorithms rely on an initial processing step that estimates a (usually sparse) set of promising points in the image. For instance, a vessel segmentation algorithm may first attempt to identify a sparse set of seed points that mostly lie on the image foreground (the vessel structures of interest). Using these seeds, the algorithm can generate a segmentation. For example, vessel tracing algorithms can be initiated from seed points. In some cases, it makes good sense to inspect the seed points and edit them prior to finalizing segmentation. This usually saves a lot of post-segmentation editing, and seeds are often much easier to edit than object outlines. In some cases a perfect set of seeds guarantees a perfect segmentation. Visit this page for more information [FARSIGHT Seed Editor]
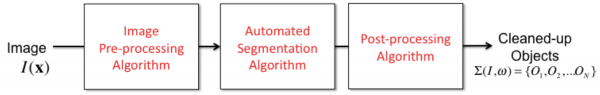
Pre-processing
This is another catch-all phrase that refers to image processing operations that precede image segmentation. Their main purpose is to help improve the results of image segmentation by "cleaning up" the image in a defined manner. For example, smoothing an image, and suppressing known image artifacts (e.g., blur, non-uniform illumination) can improve segmentation results significantly.
Post-processing
This catch-all phrase refers to operations that are conducted on the output of automated image segmentation results. Their purpose is to "clean up" the results of automated image segmentation results, and suppress/eliminate certain types of errors prior to computing any measurements of the objects. For example, if we know that the objects of interest cannot be smaller than 10 voxels in size, and we find that some of the objects delineated by automated segmentation are indeed smaller than 10 voxels, we can either delete these small objects, or look for appropriate opportunities to merge them with other objects.
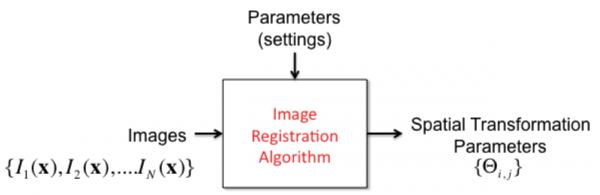
Image Registration
This is the process of spatially aligning two or more images into a common coordinate framework. Registration results in a spatial transformation function 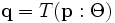 that takes a point
that takes a point 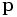 in one image to a corresponding point
in one image to a corresponding point 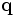 in another image. To specify the spatial transformation, we also need transformation parameters Θ. Image registration can be done for a pair of images (pairwise registration), or for a set of overlapping images at once (joint registration). The latter method is much more effective when a large number of images constituting a montage (mosaic) must be aligned together.
in another image. To specify the spatial transformation, we also need transformation parameters Θ. Image registration can be done for a pair of images (pairwise registration), or for a set of overlapping images at once (joint registration). The latter method is much more effective when a large number of images constituting a montage (mosaic) must be aligned together.
Mosaicing (Montaging)
Mosaicing (a.k.a. Montaging or Montage Synthesis) is the stitching together of multiple partial views of a region that is much larger than the field of view of the imaging instrument, into a single large seamless image of the entire region. In FARSIGHT, we are interested in many different forms of mosaicing. The simplest is to mosaic two or more grayscale images (2D/3D). We are also interested in object-level mosaicing. This happens when segmentation results for two or more overlapping image fields are stitched together to produce a segmentation mosaic.