Intrinsic Features of Blobs
From FarsightWiki
(Difference between revisions)
Dirkpadfield (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
| − | |||
These features can be calculated with two input images (Data Image and Label Image). | These features can be calculated with two input images (Data Image and Label Image). | ||
They are most commonly used for blob-like regions, such as cell nuclei. | They are most commonly used for blob-like regions, such as cell nuclei. | ||
Equations are shown for 3-dimensional space unless otherwise noted. | Equations are shown for 3-dimensional space unless otherwise noted. | ||
| + | == Glossary of Notation == | ||
| + | {| border="1px" cellpadding="3" style="text-align:left" | ||
| + | |- | ||
| + | | <math>p=(x,y,z)</math> | ||
| + | | the coordinate of a voxel (three-dimensional point in a volume image) | ||
| + | |- | ||
| + | | <math>N_p</math> | ||
| + | | a neighbor voxel of <math>p</math> | ||
| + | |- | ||
| + | | <math>l_p</math> | ||
| + | | the segmentation label at <math>p</math> | ||
| + | |- | ||
| + | | <math>I_i(p)</math> | ||
| + | | the intensity value of <math>p</math> at <math>i^{th}</math> | ||
| + | |- | ||
| + | | <math>\Omega = \{p|l_p = o\}</math> | ||
| + | | the set of voxels of an object <math>o</math> | ||
| + | |- | ||
| + | | <math>\Omega_s = \{l_p = o; \exists N_p, l_{N_p} \neq o\}</math> | ||
| + | | the set of surface voxels of the object | ||
| + | |- | ||
| + | | <math>\Omega_{in} = \Omega - \Omega_s</math> | ||
| + | | the set of interior voxels of an object | ||
| + | |- | ||
| + | | <math>\bar{p}</math> | ||
| + | | the center of mass of the object | ||
| + | |- | ||
| + | | <math>P(I)</math> | ||
| + | | normalized histogram of intensity values <math>I</math> | ||
| + | |- | ||
| + | | <math>M_{p,q,r} = \sum_{z=0}^{Z-1}\sum_{y=0}^{Y-1}\sum_{x=0}^{X-1}x^p y^q z^r I(x,y,z)</math> | ||
| + | | Raw Moment of discrete image <math>I</math> | ||
| + | |- | ||
| + | | <math>\lambda_i</math> | ||
| + | | <math>i^{th}</math> eigenvalue of covariance matrix | ||
| + | |- | ||
| + | | <math>\bar{v_i}</math> | ||
| + | | eigenvector corresponding to <math>\lambda_i</math> | ||
| + | |} | ||
| + | |||
| + | == Features == | ||
{| border="1px" cellpadding="3" style="text-align:left" | {| border="1px" cellpadding="3" style="text-align:left" | ||
|- | |- | ||
| Line 40: | Line 80: | ||
| Orientation | | Orientation | ||
| Angle between the major axis of the best-fit hyper-ellipsoid and origin. (2D) [1] | | Angle between the major axis of the best-fit hyper-ellipsoid and origin. (2D) [1] | ||
| − | | <math>tan^{-1} | + | | <math>tan^{-1}(\frac{\bar{v_1}(1)}{\bar{v_1}(0)})</math> |
|- | |- | ||
| Bounding Box Volume | | Bounding Box Volume | ||
| Line 127: | Line 167: | ||
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | ==References== | + | == References == |
[1] [http://www.insight-journal.org/browse/publication/301 itkLabelGeometryImageFilter]<br> | [1] [http://www.insight-journal.org/browse/publication/301 itkLabelGeometryImageFilter]<br> | ||
[2] [http://www.itk.org/Doxygen312/html/classitk_1_1LabelStatisticsImageFilter.html itkLabelStatisticsImageFilter]<br> | [2] [http://www.itk.org/Doxygen312/html/classitk_1_1LabelStatisticsImageFilter.html itkLabelStatisticsImageFilter]<br> | ||
Revision as of 19:02, 28 April 2009
These features can be calculated with two input images (Data Image and Label Image). They are most commonly used for blob-like regions, such as cell nuclei. Equations are shown for 3-dimensional space unless otherwise noted.
Glossary of Notation
| p = (x,y,z) | the coordinate of a voxel (three-dimensional point in a volume image) |
| Np | a neighbor voxel of p |
| lp | the segmentation label at p |
| Ii(p) | the intensity value of p at ith |
| Ω = {p | lp = o} | the set of voxels of an object o |
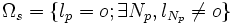
|
the set of surface voxels of the object |
| Ωin = Ω − Ωs | the set of interior voxels of an object |
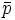
|
the center of mass of the object |
| P(I) | normalized histogram of intensity values I |
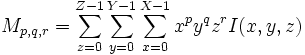
|
Raw Moment of discrete image I |
| λi | ith eigenvalue of covariance matrix |
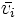
|
eigenvector corresponding to λi |
Features
| Name | Description | Formula |
| Volume | Number of voxels in the object [1] | | Ω | or M000 | {I = binary} |
| Integrated Intensity | Sum of the intensities of all voxels in the object [1] | 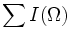 or M000 | {I = intensity} or M000 | {I = intensity}
|
| Centroid | Center of the object [1] | ![\left [ \begin{array}{ccc} \frac{M_{100}}{M_{000}}, & \frac{M_{010}}{M_{000}}, & \frac{M_{001}}{M_{000}} \end{array} \right ]|\{I=binary\}](/mw/images/math/c/0/4/c04ebc21359578e839e7056b6f67a774.png)
|
| Weighted Centroid | Uses the image intensity values to calculate the center of mass of the object [1] | ![\left [ \begin{array}{ccc} \frac{M_{100}}{M_{000}}, & \frac{M_{010}}{M_{000}}, & \frac{M_{001}}{M_{000}} \end{array} \right ]|\{I=intensity\}](/mw/images/math/0/5/c/05c7b86bceb7a3bb6f5b4d3f3fe529ef.png)
|
| Axes Lengths | The length of the axes of the ND hyper-ellipsoid fit to the object [1] | 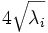
|
| Eccentricity | Ratio of the distance between the foci of the best-fit hyper-ellipsoid to the length of its major axis. (2D) [1] | 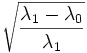
|
| Elongation | Ratio of the major axis length to minor axis length of the best-fit hyper-ellipsoid. (2D) [1] | 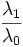
|
| Orientation | Angle between the major axis of the best-fit hyper-ellipsoid and origin. (2D) [1] | 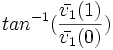
|
| Bounding Box Volume | Number of voxels in the bounding box of the object [1] | (max(X)-min(X)+1) * (max(Y)-min(Y)+1) * ... |
| Oriented Bounding Box Volume | Number of voxels in the oriented bounding box of the object. The oriented bounding box is defined as the bounding box aligned along the axes of the object. [1] | |
| Sum | Same as integrated intensity [2] | 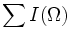 or M000 | {I = intensity} or M000 | {I = intensity}
|
| Mean | Average intensity of voxels in the object [2] | 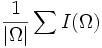
|
| Median | Middle intensity of voxels in the object [2] | |
| Minimum | Minimum intensity of voxels in the object [2] | |
| Maximum | Maximum intensity of voxels in the object [2] | |
| Sigma | Standard deviation of intensity of voxels in the object [2] | σI |
| Variance | Variance of intensity of voxels in the object [2] | 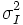
|
| Radius Variation | Standard deviation of distance from surface voxels to centroid | stddev(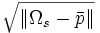
|
| Skew | Skew of the normalized intensity histogram [3] | 
|
| Energy | Energy of the normalized intensity histogram [3] | ![\sum_{I=0}^{255}[P(I)]^2](/mw/images/math/1/f/d/1fd6f9ae5d3391efdd4dcdc207610330.png)
|
| Entropy | Entropy of the normalized intensity histogram [3] | 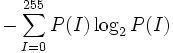
|
| Surface Gradient | Average of surface gradients | mean(G(Ωs)) |
| Interior Gradient | Average of interior gradients | mean(G(Ωin)) |
| Interior Intensity | Average of interior intensities | mean(I(Ωin)) |
| Surface Intensity | Average of surface intensities | mean(I(Ωs)) |
| Intensity Ratio | Ratio of surface intensity to interior intensity | 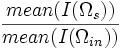
|
| Shared Boundary | Ratio of object "edges" that touch another object to total number of object "edges | |
| Surface Area | Number of voxels on surface of the object [4] | | Ωs | |
| Shape | Ratio of surface voxels to total voxels - compactness or thinness of object [5] | 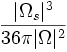
|
References
[1] itkLabelGeometryImageFilter
[2] itkLabelStatisticsImageFilter
[3] Umbaugh, S. E., Y.-S. Wei, et al. (1997). "Feature extraction in image analysis. A program for facilitating data reduction in medical image classification." Engineering in Medicine and Biology Magazine, IEEE 16(4): 62-73.
[4] Lohmann, G. (1998). Volumetric Image Analysis, Wiley
[5] Theodoridis, S. and K. Koutroumbas (1999). Pattern recognition. San Diego, Academic Press.
[6] Kitware Source Newsletter