Nuclear Segmentation
| Line 7: | Line 7: | ||
| − | [[Image:Nuc_seg_flow_chart.png | | + | [[Image:Nuc_seg_flow_chart.png |thumb|600px|center]] |
Revision as of 18:00, 18 April 2009
This page provides a brief overview of the nuclear segmentation algorithm, usage instruction, and sample results.
Overview of the Algorithm
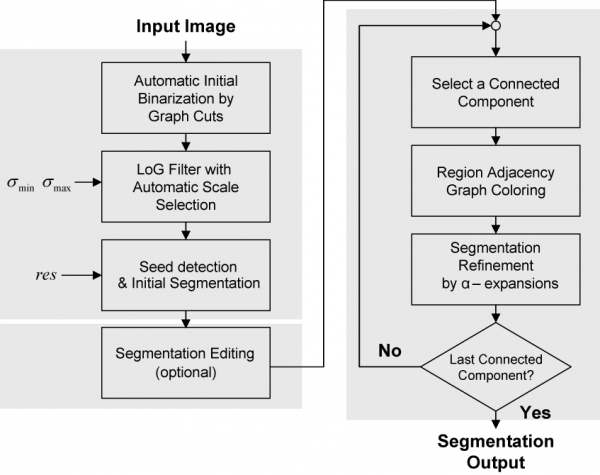
Automatic segmentation of cell nuclei is a widely needed essential step several biological applications. Although much progress has been made, this step continues to require improvements in accuracy, speed, level of automation, and ease of adaptability to new applications.
The segmentation process starts by binarizing the image based on the Graph-Cuts algorithm with automatic learning using minimum error thresholding. In the second step, the nuclear channel image is transformed into another one in which nuclei are represented by blobs of which local maxima points are used to represent seed points of nuclei. The transformation is achieved using a multi-scale Laplacian of Gaussian (LoG) filter, with the addition of automatic and adaptive scale selection. Given a set of seed points and the output of the LoG filter, the next step is to delineate each nucleus. For this, we use a fast clustering technique, called local maximum clustering. In the last step, nuclear contours are refined using a multi-label graph-cuts approach with alpha-expansions. In order to reduce memory requirements and processing time, alpha-expansions is preceded by a graph-coloring step that permits processing a large number of nuclei in parallel. Figure 1 shows a flow-chart illustrating the different steps of our work.