STrenD: Subspace Trend Discovery
Contents |
Software Interface
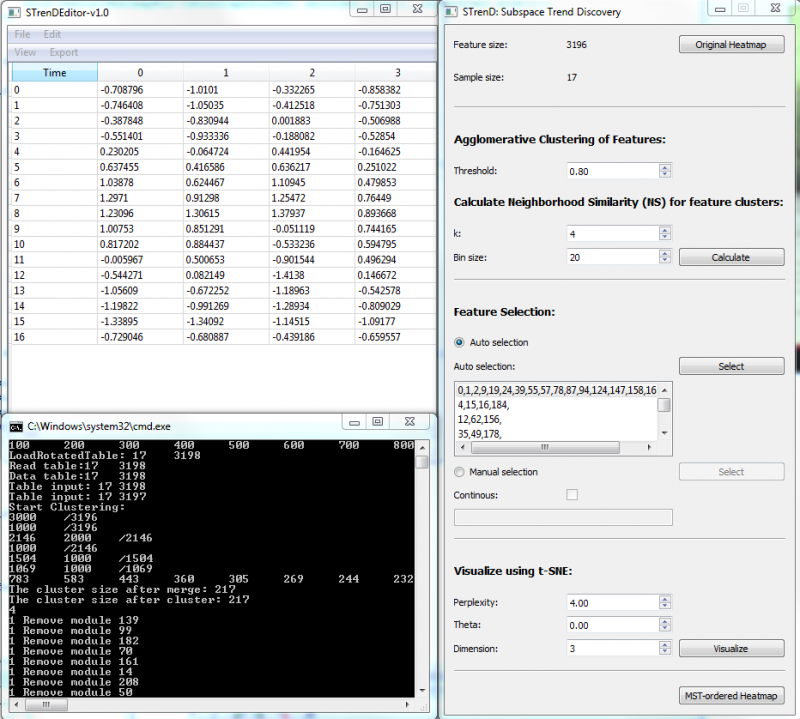
1. Load Tab-delimited txt file. If columns are features and rows are samples, File/Load Table; If columns are samples and rows are features, File/Load Rotated Table;
2. Calculate for feature clustering and pair-wise neighborhood similarity (NS);
3. Auto selection: push select for automatic thresholding on NS matrix to provide a list of non-overlapping feature subsets (size >=3). The largest subset, on top of the list, is selected by default;
4. Manual selection: push select to visualize co-clustered NS matrix and select a group of features that have high NS values by left clicking on the top-left starting square and releasing on the right-bottom ending square. The user can also input feature cluster index in the editor, separated by comma; The old selection is kept when Continuous is checked, or else the old selection is erased.
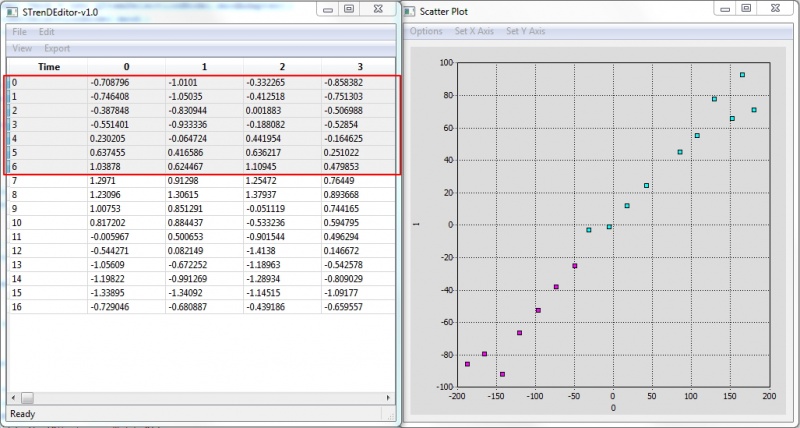
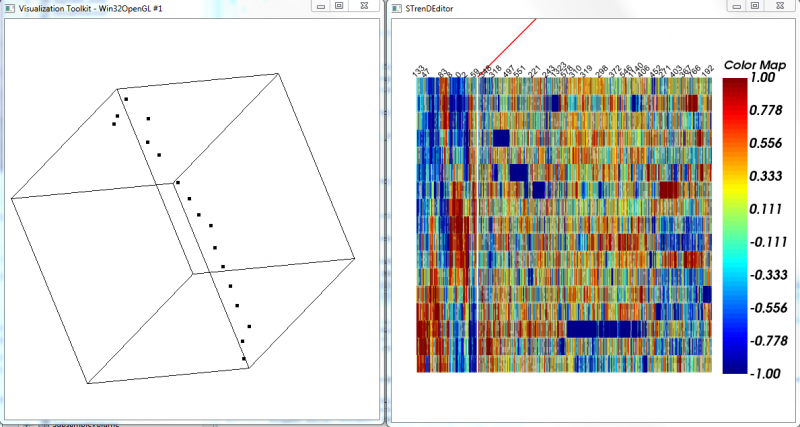
5. Visualize to provide a 2-D or 3-D visualization using t-SNE("dimension" higher than 3 would be visualized in 2D with a selected pair of dimensions);
6. MST-ordered Heatmap to visualize a heatmap with rows arranged by the depth-first order of MST on selected data and columns arranged by a hierarchical clustering of features. The selected ones are separated by a red line from the rest.
Download
STrenD-v1.0 (implemented in C++) is available to download at
Windows 64 bit:
Release: https://github.com/YanXuHappygela/STrenD-release-1.0
Source codes: https://github.com/YanXuHappygela/STrenD-source-1.0
Matlab wrapper is coming up soon!
If you have any problem with the software, please report to yansoftwareus@gmail.com. Thank you.
Test on Cell Cycle Microarray data
For test dataset "cellCycleMicroarray.txt" with default param settings:
File/Load Rotated Table (cellCycleMicroarray.txt) -> Auto selection:select ->Visualize->MST-ordered Heatmap
Actively-linked Visualization
Output Files
For test dataset "cellCycleMicroarray.txt" with 17 samples of 3196 dimensions, clustering sigma = 0.8, k = 4:
1. 3196_17_0.8_clustering.txt: agglomerative clustering result, containing index and feature names;
2. 3196_17_0.8_4_NS.txt: pair-wise neighborhood similarity matrix of feature clusters;
3. Shanbhag.txt: intermediate outputs for Shanbhag thresholding;
4. 3196_17_0.8_4_AutoSelFeatures.txt: selected feature index and names;
5. data_selected_vis.txt: table of normalized data with selected features for visualization;
6. vis_coordinates.txt: output coordinates for visualization after dimension reduction by t-SNE.