FARSIGHT Framework
We start by assuming that we are given one or more multi-dimensional image(s) I(x,y,z,λ,t), as appropriate. The hardest first step to automated image analysis is image segmentation. We simplify this task using a 'divide and conquer' strategy. We recognize that some of the best-available segmentation algorithms are model based. Usually they require a model describing the expected geometry of biological objects, and a model describing the imaging process and expected defects such as noise and artifacts.
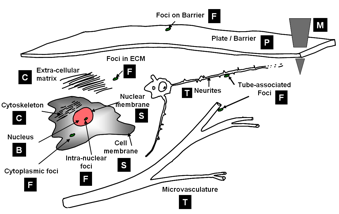
Computational taxonomy of Object Morphologies: We are primarily interested in fluorescence microscopy at the cell and tissue level (0.1 − 1000μm). Within this realm, we have found that, in spite of the variability in biological forms, it is possible to identify a “short list” of frequently occurring morphologies of biological entities – blobs (B), tubes (T), shells (S), foci (F), plates (P), clouds (C), and man-made objects (M). These morphologies are illustrated in the following figure:
Short list of object morphologies frequently observed in fluorescence cell / tissue imagery: B = Blobs; T = Tubes; S = Shells; F = Foci/ punctae; P = Plates; C = Clouds; M = Man-made objects.
Commonly, blobs correspond to cell nuclei, tubes correspond to neuron processes/vasculature, shells correspond to nuclear/cell membranes, foci represent localized molecular concentrations such as mRNA at the site of transcription, cell-cell adhesions and synapses, plates correspond to basal laminae, clouds correspond to cytoplasmic markers, and man-made objects correspond to implanted devices. We require the user to designate what morphological category a given biological object corresponds to. This information specifies a segmentation algorithm from our library of segmentation algorithms.