FARSIGHT Framework
The following paragraphs provide a brief overview of the flow of steps underlying FARSIGHT.
Starting Point: We start by assuming that we are given one or more multi-dimensional image(s) I(x,y,z,λ,t), as appropriate. The image data and its associated metadata can be stored in a folder on a computer's secondary storage (disk), or in an image database such as the [Open Microscopy Environment] consortium's OMERO system. There are a large number of image file formats in use. FARSIGHT uses the [OME Bioformats ] module to help translate file formats and manage the all-important metadata.
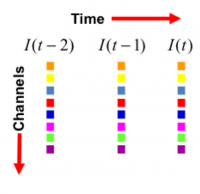
Data Unmixing: The hardest first step to automated image analysis is image segmentation. We simplify this task using a 'divide and conquer' strategy. We recognize that some of the best-available segmentation algorithms are model based. Usually they require a model describing the expected geometry of biological objects, and a model describing the imaging process and expected defects such as noise and artifacts. Modern microscopes have highly developed spectral imaging hardware and software systems for spectral unmixing. They are very effective at breaking down the image data into a set of non-overlapping channels. When this procedure works correctly, each channel only contains one type of biological object - we call them "pure channels". This greatly simplifies the task of segmentation by enabling our divide and conquer segmentation strategy (more on this below). The figure below shows an easy way to picture a multi-dimensional image that has been split into a set of channels (each row corresponds to a channel) and temporal sampling points (each column represents a point in time). If we have a parallel computer available, it can be made to process all of these tiles (representing a 2-D or 3-D dataset) in parallel.
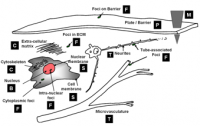
Computational taxonomy of Object Morphologies: We are primarily interested in fluorescence microscopy at the cell and tissue level (0.1 − 1000μm). Within this realm, we have found that, in spite of the variability in biological forms, it is possible to identify a “short list” of frequently occurring morphologies of biological entities – blobs (B), tubes (T), shells (S), foci (F), plates (P), clouds (C), and man-made objects (M). These morphologies are illustrated in the following figure:
Commonly, blobs correspond to cell nuclei, tubes correspond to neuron processes/vasculature, shells correspond to nuclear/cell membranes, foci represent localized molecular concentrations such as mRNA at the site of transcription, cell-cell adhesions and synapses, plates correspond to basal laminae, clouds correspond to cytoplasmic markers, and man-made objects correspond to implanted devices.
Which class should I choose? We require the user to designate what morphological category a given biological object corresponds to. This information specifies a segmentation algorithm from our library of segmentation algorithms. Each algorithm is specialized for the corresponding morphological category. The morphological class of an image object can sometimes be interpreted in more than one manner, suggesting different segmentation algorithm choices. For instance, as foci become larger, it may be advantageous to interpret them as blobs instead. The optimal choice is the one that leads to the lowest segmentation error. Specialized algorithms can achieve higher levels of automation, accuracy, speed, and robustness.
The Systems Biology Connection: Aside from its morphology, each biological object has a biophysical categorization from a systems biology standpoint. From a quantitation standpoint, these objects represent one of four types of biological entities: (i) a volumetric compartment; (ii) a surface with negligible volume; or (iii) a functional signal that could be distributed over compartments/ surfaces/ curves.
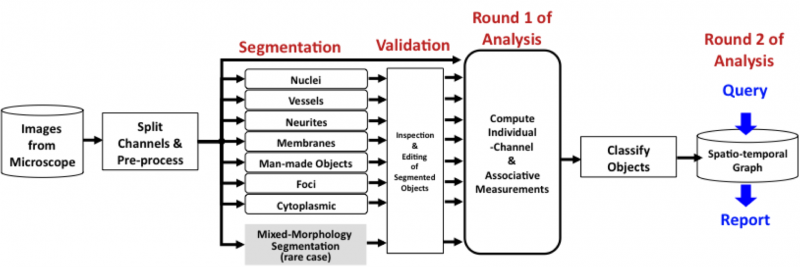
Dealing with an imperfect segmentation: Even the best-available segmentation algorithms have a non-zero error rate. In order to apply them on a large scale to complex problems, it is essential to ensure that: (i) they are run with optimal parameter settings; and (ii) their outputs are inspected and validated before computing the quantitative measurements of eventual interest. With this in mind, the FARSIGHT system will incorporate two innovative software tools. The rapid prototyping system (RPS) is our computer-aided tool for seeking out the best parameter settings, and our edit-based validation system (EVS) is a tool for efficient inspection and corrective editing of segmentations.
Computing Measurements - Round 1: Once the user is satisfied with the segmentation results, he/she can compute diverse measurements (features) of objects in the image and their associations. The diversity of biological investigational objectives implies that different measurements are important for different studies. Nevertheless, the types of measurements of interest fall into two broad categories: (i) intrinsic; and (ii) associative. Intrinsic measurements quantify the features of objects in a single channel. Associative measurements, on the other hand, quantify relationships between objects from one or more channels. We are interested in spatial associations as well as associations over time (e.g., tracking assignments). Most of these features are intended for use by the biologist. In addition to these "end use" features, we also compute some "diagnostic features" whose purpose is to help diagnose segmentation/classification errors.
Object Classification: Using intrinsic and associative features, we are able to classify objects in the image (usually cell types). For this, the FARSIGHT toolkit provides a library of pattern analysis algorithms. In some applications, the analysis ends with object classification. In others, it sets the stage for further analysis, as described below.
Computing Measurements - Round 2: Once all biological objects have been segmented and categorized, a fresh round of quantitative analysis becomes possible. For this, we rely on extensive use of graph theory. We start by representing our collection of objects and associations as an attributed graph data structure. These graphs can be visualized and edited as needed. We next cast biological questions as queries on the attributed graph. Summaries of the data are generated by FARSIGHT in response to these queries. This is our [Tissue Nets] module. We expect to implement increasingly more sophisticated queries in the future.
FARSIGHT Image Analysis Pipeline: The FARSIGHT associative image analysis pipeline is formed by stringing together the above steps (as needed) in the form of a script written in the open source [Python language]. This language is easy to learn and extremely robust. It has been used for some of the largest computational projects and we like it too.