FARSIGHT Framework
| Line 1: | Line 1: | ||
'''Starting Point: '''We start by assuming that we are given one or more multi-dimensional image(s) <math>I(x,y,z,\lambda,t)</math>, as appropriate. The hardest first step to automated image analysis is image segmentation. We simplify this task using a 'divide and conquer' strategy. We recognize that some of the best-available segmentation algorithms are model based. Usually they require a model describing the expected geometry of biological objects, and a model describing the imaging process and expected defects such as noise and artifacts. | '''Starting Point: '''We start by assuming that we are given one or more multi-dimensional image(s) <math>I(x,y,z,\lambda,t)</math>, as appropriate. The hardest first step to automated image analysis is image segmentation. We simplify this task using a 'divide and conquer' strategy. We recognize that some of the best-available segmentation algorithms are model based. Usually they require a model describing the expected geometry of biological objects, and a model describing the imaging process and expected defects such as noise and artifacts. | ||
| + | |||
| + | '''Data Unmixing: ''' Modern microscopes have highly developed spectral imaging hardware and software systems for spectral unmixing. They are very effective at breaking down the image data into a set of non-overlapping '''channels'''. In most cases, each channel only contains one type of biological object. This greatly simplifies the task of segmentation by enabling our divide and conquer segmentation strategy (more on this below). | ||
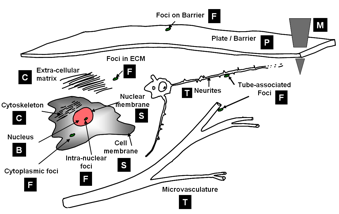
'''Computational taxonomy of Object Morphologies: '''We are primarily interested in fluorescence microscopy at the cell and tissue level <math>(0.1 - 1000 \mu m)</math>. Within this realm, we have found that, in spite of the variability in biological forms, it is possible to identify a “short list” of frequently occurring morphologies of biological entities – blobs (B), tubes (T), shells (S), foci (F), plates (P), clouds (C), and man-made objects (M). These morphologies are illustrated in the following figure: | '''Computational taxonomy of Object Morphologies: '''We are primarily interested in fluorescence microscopy at the cell and tissue level <math>(0.1 - 1000 \mu m)</math>. Within this realm, we have found that, in spite of the variability in biological forms, it is possible to identify a “short list” of frequently occurring morphologies of biological entities – blobs (B), tubes (T), shells (S), foci (F), plates (P), clouds (C), and man-made objects (M). These morphologies are illustrated in the following figure: | ||
| Line 7: | Line 9: | ||
Short list of object morphologies frequently observed in fluorescence cell / tissue imagery: B = Blobs; T = Tubes; S = Shells; F = Foci/ punctae; P = Plates; C = Clouds; M = Man-made objects. | Short list of object morphologies frequently observed in fluorescence cell / tissue imagery: B = Blobs; T = Tubes; S = Shells; F = Foci/ punctae; P = Plates; C = Clouds; M = Man-made objects. | ||
| − | Commonly, blobs correspond to cell nuclei, tubes correspond to neuron processes/vasculature, shells correspond to nuclear/cell membranes, foci represent localized molecular concentrations such as mRNA at the site of transcription, cell-cell adhesions and synapses, plates correspond to basal laminae, clouds correspond to cytoplasmic markers, and man-made objects correspond to implanted devices. We require the user to designate what morphological category a given biological object corresponds to. This information specifies a segmentation algorithm from our [[library of segmentation algorithms]]. Each algorithm is specialized for the corresponding morphological category. | + | Commonly, blobs correspond to cell nuclei, tubes correspond to neuron processes/vasculature, shells correspond to nuclear/cell membranes, foci represent localized molecular concentrations such as mRNA at the site of transcription, cell-cell adhesions and synapses, plates correspond to basal laminae, clouds correspond to cytoplasmic markers, and man-made objects correspond to implanted devices. |
| + | |||
| + | '''Which class should I choose? '''We require the user to designate what morphological category a given biological object corresponds to. This information specifies a segmentation algorithm from our [[library of segmentation algorithms]]. Each algorithm is specialized for the corresponding morphological category. The morphological class of an image object can sometimes be interpreted in more than one manner, suggesting different segmentation algorithm choices. For instance, as foci become larger, it may be advantageous to interpret them as blobs instead. The optimal choice is the one that leads to the lowest segmentation error. Specialized algorithms can achieve higher levels of automation, accuracy, speed, and robustness. | ||
'''FARSIGHT Image Analysis Pipeline: ''' | '''FARSIGHT Image Analysis Pipeline: ''' | ||
Revision as of 20:18, 9 April 2009
Starting Point: We start by assuming that we are given one or more multi-dimensional image(s) I(x,y,z,λ,t), as appropriate. The hardest first step to automated image analysis is image segmentation. We simplify this task using a 'divide and conquer' strategy. We recognize that some of the best-available segmentation algorithms are model based. Usually they require a model describing the expected geometry of biological objects, and a model describing the imaging process and expected defects such as noise and artifacts.
Data Unmixing: Modern microscopes have highly developed spectral imaging hardware and software systems for spectral unmixing. They are very effective at breaking down the image data into a set of non-overlapping channels. In most cases, each channel only contains one type of biological object. This greatly simplifies the task of segmentation by enabling our divide and conquer segmentation strategy (more on this below).
Computational taxonomy of Object Morphologies: We are primarily interested in fluorescence microscopy at the cell and tissue level (0.1 − 1000μm). Within this realm, we have found that, in spite of the variability in biological forms, it is possible to identify a “short list” of frequently occurring morphologies of biological entities – blobs (B), tubes (T), shells (S), foci (F), plates (P), clouds (C), and man-made objects (M). These morphologies are illustrated in the following figure:
Short list of object morphologies frequently observed in fluorescence cell / tissue imagery: B = Blobs; T = Tubes; S = Shells; F = Foci/ punctae; P = Plates; C = Clouds; M = Man-made objects.
Commonly, blobs correspond to cell nuclei, tubes correspond to neuron processes/vasculature, shells correspond to nuclear/cell membranes, foci represent localized molecular concentrations such as mRNA at the site of transcription, cell-cell adhesions and synapses, plates correspond to basal laminae, clouds correspond to cytoplasmic markers, and man-made objects correspond to implanted devices.
Which class should I choose? We require the user to designate what morphological category a given biological object corresponds to. This information specifies a segmentation algorithm from our library of segmentation algorithms. Each algorithm is specialized for the corresponding morphological category. The morphological class of an image object can sometimes be interpreted in more than one manner, suggesting different segmentation algorithm choices. For instance, as foci become larger, it may be advantageous to interpret them as blobs instead. The optimal choice is the one that leads to the lowest segmentation error. Specialized algorithms can achieve higher levels of automation, accuracy, speed, and robustness.
FARSIGHT Image Analysis Pipeline: