Tracking
(→Unmixing) |
(→Unmixing) |
||
| Line 23: | Line 23: | ||
| − | [[Image:Unmixing.jpg|500px|thumb|(left)put in the caption]] | + | [[Image:Unmixing.jpg|500px|thumb|(left)]put in the caption]] |
| − | + | ||
| − | + | ||
| − | + | ||
===Segmentation=== | ===Segmentation=== | ||
Revision as of 17:55, 9 June 2009
This page describes our main methodology in doing a 5-D Image analysis.
Contents |
5-D Image Analysis
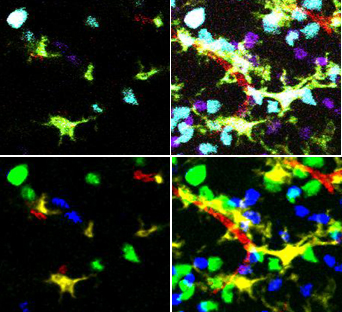
5-D Image analysis involves object extraction and tracking of multi-channel data. A typical dataset is shown on the right, with two types of T-cells that need to be tracked. It also has dendritic cells and blood vessels imaged in the same spatial context. The main framework for analyzing such a movie has been described in FARSIGHT_Framework. In a nut shell, we adopt a divide and conquer strategy in segmenting the different channels. We then track each channel individually and computer both intrinsic features, which are intrinsic to an object and associative features between an object and other channels.
Tracking Algorithm
Our approach to tracking involves multiple steps. They are
- Unmix the channels
- Segment each channel independently
- Track each channel independently
- Compute intrinsic features of each channel and associative features between channels
Unmixing
We have implemented a simple unmixing algorithm that involves a pre-processing step of median filtering on each mixed channel using a 3x3x3 kernel followed by a non-maximum suppression of the channels.
The unmixing itself is really classification. The values of corresponding pixels in the different color channels are compared and only the largest value is maintained The corresponding pixels in the other channels are set to zero.
Segmentation
Segmentation of each channel is computed independent of the remaining channels through a custom tailored algorithm depending on the object morphology present in the channel. We use a nuclear segmentation program for segmenting blob like cell nuclei. We use a combination of thresholding, binary morphological operations to segment dendritic cells. Blood vessel are segmented by first binarizing them followed by a skeletonization algorithm to extract their centerlines.
TODO:Add sample segmentation outputs
Tracking
Tracking algorithm tracks each channel separately based on their segmentation. Tracking computes the optimum time association between objects of two consecutive time points based on multiple hypotheses. It uses the Hungarian algorithm to find the optimum association based on a matrix of cost functions.
TODO: Add more text describing the tracking algorithm. Perhaps add illustrations.
Feature computation
FARSIGHT computes a rich set of features for objects. Apart from the intrinsic features, we also compute associative features between objects of different channels. In case of 5-D image analysis, we also compute time based features.
TODO: Add time based features to Features
Tracking Program
Usage:
track
Input parameters are contained in the file filename.conf located in the same directory of execution. It contains one row per 3-D image to be processed of the form
DatasetID "Filename" channel_id time_point
An example file look like the following lines
dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w1_t10.tif" 1 10 dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w2_t10.tif" 2 10 dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w3_t10.tif" 3 10 dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w4_t10.tif" 4 10 dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w1_t11.tif" 1 11 dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w2_t11.tif" 2 11 dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w3_t11.tif" 3 11 ... dataset1 "data/wF5p120307m1s5-t10/wF5p120307m1s5_w4_t14.tif" 4 14
First line represents one 3-D image of a movie named "dataset1" with filename "data/wF5p120307m1s5-t10/wF5p120307m1s5_w1_t10.tif". This file is from channel 1 and time point 10.
Output Files: The output files for the program are of 4 types.
- Unmixed files : Contains the individual channels after running our unmixing algorithm.
- Segmented files: Contains 16-bit images of the output of segmentation algorithm. Each cell in a time point has a unique index.
- Tracking output files: Contains 16-bit images of the output of tracking algorithm. Each cell in a time point has the index of its track.
- Feature files: Contains intrinsic and associative features computed after the tracking step.
Tracking Editor
Authors
The following people are involved in this project