Tracking
(→Tracking Program) |
|||
| (41 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| + | This page describes our main methodology in doing a 5-D Image analysis. | ||
| + | |||
==5-D Image Analysis== | ==5-D Image Analysis== | ||
| − | + | 5-D Image analysis involves object extraction and tracking of multi-channel 3-D time-lapse image data of living specimens. For a description of how these are acquired, see this protocols page [[Imaging_Protocols]]. These movies reveal the rich and active environment in a developing mouse thymus - an organ that plays a critical role in the developing immune system. | |
| − | 5-D Image analysis involves object extraction and tracking of | + | |
| + | A sample 4-color, 5-D dataset is shown below, with two types of thymocytes that need to be tracked. It also has dendritic cells and blood vessels imaged in the same spatial context. The strategy for analyzing such a movie has been described in [[FARSIGHT_Framework]]. In a nutshell, we use an unmixing algorithm to split the data into a set of non-overlapping channels. This allows us to adopt a divide and conquer strategy in segmenting the cells and vasculature in the different channels. We then track each cell (one channel at a time) individually. The results of segmentation and tracking allows us to compute both [[Features|intrinsic features]] for each cell using the FARSIGHT feature computation library, and [[Features|associative features]] between objects (within and across channels). Basically, we think of tracking correspondences as temporal associations. | ||
| + | |||
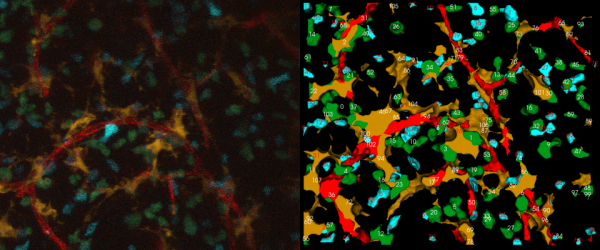
| + | [[Image:5D_sample_ena.png|600px|thumb|center|(left)A sample 2-D projection of a single time snapshot from a 5-D movie from the immune system. (right)The corresponding tracking output (red-blood vessels, orange- dendritic cells, blue-T cells with Cyan Fluorescent Protein, green-T cells with Green Fluorescent Protein)]] | ||
| + | |||
| + | == Cell Tracking Algorithm == | ||
| + | |||
| + | Our approach to tracking involves multiple steps. They are | ||
| + | |||
| + | *Unmix the channels | ||
| + | *Segment each channel independently | ||
| + | *Track each channel independently | ||
| + | *Compute intrinsic features of each channel and associative features between channels | ||
| + | |||
| + | ===Spectral Unmixing=== | ||
| + | |||
| + | In these datasets, the fluorescence emission is detected by a small set of (4) photomultiplers (PMTs), rather than an array of detectors that can collect a finely resolved spectrum at each voxel. Consequently, the spectral unmixing problem must be approached with limited data in mind. For this, we have implemented a simple unmixing algorithm that involves a pre-processing step of median filtering on each mixed channel using a 3x3x3 kernel followed by a non-maximum suppression of the channels. | ||
| + | |||
| + | This type of unmixing can be thought of as a kind ofvoxel classification. The values of corresponding pixels in the different color channels are compared and only the largest value is kept. The corresponding pixels in the other channels are set to zero. This method serves our need to delineate each cell and vessel segment. | ||
| + | |||
| + | |||
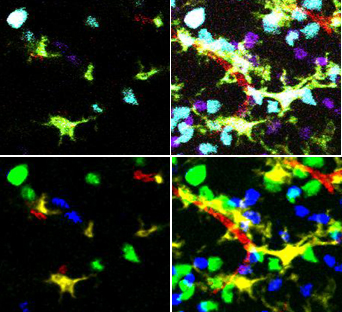
| + | [[Image:Unmixing.jpg|500px|thumb|'''After unmixing, classification of cell types is simple.'''<br /> | ||
| + | (top left) single plane from the original microscope image <br /> | ||
| + | (top right) Z projection of the original microscope images<br /> | ||
| + | (bottom left) unmixed single plane<br /> | ||
| + | (bottom right) unmixed projection]] | ||
| + | |||
| + | ===Segmentation=== | ||
| + | |||
| + | Segmentation of each channel is computed independent of the remaining channels through a custom tailored algorithm depending on the object morphology present in the channel. We use a [[Nuclear Segmentation|nuclear segmentation]] program for segmenting blob like cell nuclei. We use a combination of thresholding, binary morphological operations to segment dendritic cells. Blood vessel are segmented by first binarizing them followed by a skeletonization algorithm to extract their centerlines. | ||
| + | |||
| + | |||
| + | |||
| + | |||
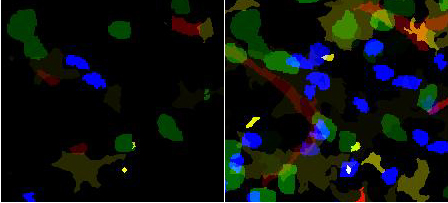
| + | [[Image:Labeled-segmented_4_color-pair.jpg|500px|thumb|'''Segmentation of all channels'''<br /> | ||
| + | (left) A single '''Z''' plane. The identification of different segmented objects is shown as different intensities. | ||
| + | (right) Z projection.<br /> | ||
| + | blue and green- two populations of T cells , yellow/brown- dendritic cells, red- blood vessels]] | ||
| + | |||
| + | ===Tracking=== | ||
| + | |||
| + | Tracking algorithm tracks each channel separately based on their segmentation. Tracking computes the optimum time association between objects of two consecutive time points based on multiple hypotheses. It uses the Hungarian algorithm to find the optimum association based on a matrix of cost functions. | ||
| + | |||
| + | '''TODO: Add more text describing the tracking algorithm. Perhaps add illustrations.''' | ||
| + | |||
| + | |||
| + | ===Feature computation=== | ||
| + | |||
| + | FARSIGHT computes a rich set of [[Features|features]] for objects. Apart from the [[Intrinsic_Features_of_Blobs|intrinsic features]], we also compute associative features between objects of different channels. In case of 5-D image analysis, we also compute time based features. | ||
| + | |||
| + | ''' TODO: Add time based features to [[Features]] ''' | ||
== Tracking Program == | == Tracking Program == | ||
| − | + | The code for tracking can be obtained from the Farsight repository. The input data folder, the current working directory and the directory containing the executables for the program are set by assigning the variables ''"data_directory","cwd" and "exe_dir"'' in the python script, ''"tracker.py"''. | |
| − | + | ||
| − | + | For example: | |
| − | + | ||
| + | data_directory = 'C:\\Python31\\complete tracker\\bin files\\release\\data\\wF5p120307m1s5-t10' | ||
| + | cwd = 'C:\\Python31\\complete tracker\\bin files\\release' | ||
| + | exe_dir = 'C:\\Python31\\complete tracker\\bin files\\release' | ||
| − | + | The input data is moved to the ''"data_directory"'' folder and the output images are obtained in the ''"\cache"'' folder. | |
| + | To run the python script, give in the command: ''python tracker.py'' at the commandline after changing the path to the python folder in your system if the path is not set already. | ||
| − | + | The python script has four segments: | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | 1. Unmixing: | |
| + | The images from the ''"data directory"'' are taken as input by the program and the unmixed files are written to the ''\cache'' folder. | ||
| + | |||
| + | 2. Segmentation: | ||
| + | The unmixed images from the ''\cache'' folder are taken as the input for the segmentation module. There is a choice between two kinds of segmentation, i.e. the user can choose to segment the unmixed files using yousef segmentation or a simple binary morphological operators based segmentation. The name of the configuration file for the nuclei segmentation is assigned to the variable, ''"nuclei_segmentation_cfg"''. | ||
| + | The ''vessel tracing'' is done for the channel which has vessels. | ||
| + | |||
| + | 3.Tracking: | ||
| + | The segmented files are taken as input for the Tracking module from the ''\cache'' folder. The channels numbers for the channels to be tracked are added to the variable ''"channels_to_track"''. The output images are written to the ''\cache'' folder. | ||
| + | |||
| + | 4. Feature Computation: | ||
| + | The features of some channels are compared against other channels and are written into two files respectively. The output text files are written to the ''\cache'' folder. | ||
| + | |||
| + | '''Output Files''': The output files for the program are of 4 types. | ||
| + | |||
| + | *Unmixed files : Contains the individual channels after running our unmixing algorithm. | ||
| + | *Segmented files: Contains 16-bit images of the output of segmentation algorithm. Each cell in a time point has a unique index. | ||
| + | *Tracking output files: Contains 16-bit images of the output of tracking algorithm. Each cell in a time point has the index of its track. | ||
| + | *Feature files: Contains intrinsic and associative features computed after the tracking step. | ||
== Tracking Editor == | == Tracking Editor == | ||
| + | |||
| + | ==Authors== | ||
| + | |||
| + | The following people are involved in this project | ||
| + | |||
| + | *[[User:Arun|Arunachalam Narayanaswamy]] | ||
| + | *[[User:Ying|Ying Chen]] | ||
| + | *[[User:Ena|Ena Ladi]] | ||
| + | *[[User:Paul|Paul Herzmark]] | ||
Latest revision as of 21:26, 3 August 2009
This page describes our main methodology in doing a 5-D Image analysis.
Contents |
5-D Image Analysis
5-D Image analysis involves object extraction and tracking of multi-channel 3-D time-lapse image data of living specimens. For a description of how these are acquired, see this protocols page Imaging_Protocols. These movies reveal the rich and active environment in a developing mouse thymus - an organ that plays a critical role in the developing immune system.
A sample 4-color, 5-D dataset is shown below, with two types of thymocytes that need to be tracked. It also has dendritic cells and blood vessels imaged in the same spatial context. The strategy for analyzing such a movie has been described in FARSIGHT_Framework. In a nutshell, we use an unmixing algorithm to split the data into a set of non-overlapping channels. This allows us to adopt a divide and conquer strategy in segmenting the cells and vasculature in the different channels. We then track each cell (one channel at a time) individually. The results of segmentation and tracking allows us to compute both intrinsic features for each cell using the FARSIGHT feature computation library, and associative features between objects (within and across channels). Basically, we think of tracking correspondences as temporal associations.
Cell Tracking Algorithm
Our approach to tracking involves multiple steps. They are
- Unmix the channels
- Segment each channel independently
- Track each channel independently
- Compute intrinsic features of each channel and associative features between channels
Spectral Unmixing
In these datasets, the fluorescence emission is detected by a small set of (4) photomultiplers (PMTs), rather than an array of detectors that can collect a finely resolved spectrum at each voxel. Consequently, the spectral unmixing problem must be approached with limited data in mind. For this, we have implemented a simple unmixing algorithm that involves a pre-processing step of median filtering on each mixed channel using a 3x3x3 kernel followed by a non-maximum suppression of the channels.
This type of unmixing can be thought of as a kind ofvoxel classification. The values of corresponding pixels in the different color channels are compared and only the largest value is kept. The corresponding pixels in the other channels are set to zero. This method serves our need to delineate each cell and vessel segment.
Segmentation
Segmentation of each channel is computed independent of the remaining channels through a custom tailored algorithm depending on the object morphology present in the channel. We use a nuclear segmentation program for segmenting blob like cell nuclei. We use a combination of thresholding, binary morphological operations to segment dendritic cells. Blood vessel are segmented by first binarizing them followed by a skeletonization algorithm to extract their centerlines.
Tracking
Tracking algorithm tracks each channel separately based on their segmentation. Tracking computes the optimum time association between objects of two consecutive time points based on multiple hypotheses. It uses the Hungarian algorithm to find the optimum association based on a matrix of cost functions.
TODO: Add more text describing the tracking algorithm. Perhaps add illustrations.
Feature computation
FARSIGHT computes a rich set of features for objects. Apart from the intrinsic features, we also compute associative features between objects of different channels. In case of 5-D image analysis, we also compute time based features.
TODO: Add time based features to Features
Tracking Program
The code for tracking can be obtained from the Farsight repository. The input data folder, the current working directory and the directory containing the executables for the program are set by assigning the variables "data_directory","cwd" and "exe_dir" in the python script, "tracker.py".
For example:
data_directory = 'C:\\Python31\\complete tracker\\bin files\\release\\data\\wF5p120307m1s5-t10' cwd = 'C:\\Python31\\complete tracker\\bin files\\release' exe_dir = 'C:\\Python31\\complete tracker\\bin files\\release'
The input data is moved to the "data_directory" folder and the output images are obtained in the "\cache" folder. To run the python script, give in the command: python tracker.py at the commandline after changing the path to the python folder in your system if the path is not set already.
The python script has four segments:
1. Unmixing: The images from the "data directory" are taken as input by the program and the unmixed files are written to the \cache folder.
2. Segmentation: The unmixed images from the \cache folder are taken as the input for the segmentation module. There is a choice between two kinds of segmentation, i.e. the user can choose to segment the unmixed files using yousef segmentation or a simple binary morphological operators based segmentation. The name of the configuration file for the nuclei segmentation is assigned to the variable, "nuclei_segmentation_cfg". The vessel tracing is done for the channel which has vessels.
3.Tracking: The segmented files are taken as input for the Tracking module from the \cache folder. The channels numbers for the channels to be tracked are added to the variable "channels_to_track". The output images are written to the \cache folder.
4. Feature Computation: The features of some channels are compared against other channels and are written into two files respectively. The output text files are written to the \cache folder.
Output Files: The output files for the program are of 4 types.
- Unmixed files : Contains the individual channels after running our unmixing algorithm.
- Segmented files: Contains 16-bit images of the output of segmentation algorithm. Each cell in a time point has a unique index.
- Tracking output files: Contains 16-bit images of the output of tracking algorithm. Each cell in a time point has the index of its track.
- Feature files: Contains intrinsic and associative features computed after the tracking step.
Tracking Editor
Authors
The following people are involved in this project