Trace Editor/Features
From FarsightWiki
(Difference between revisions)
(→Algorithms) |
|||
| Line 1: | Line 1: | ||
| − | + | ='''Features and algorithms for [[Trace Editor]]'''= | |
| − | ='''Feature List'''= | + | =='''Feature List'''== |
{| border="1px" cellpadding="2" style="text-align:left" | {| border="1px" cellpadding="2" style="text-align:left" | ||
| Line 53: | Line 53: | ||
|} | |} | ||
| − | ='''Algorithms'''= | + | =='''Algorithms'''== |
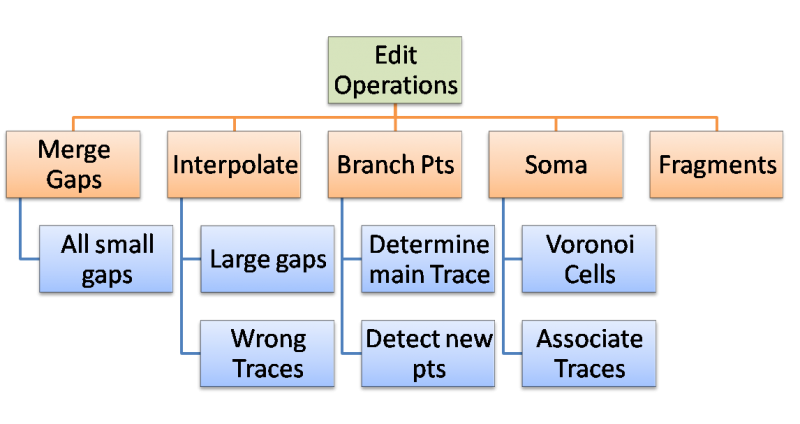
The algorithms suggested are used to control the editing process allowing for rule based cluster editing. The Goal is to complete group editing in five steps. | The algorithms suggested are used to control the editing process allowing for rule based cluster editing. The Goal is to complete group editing in five steps. | ||
[[Image:Edit_Operations.png|800px|thumb|center|Proposed steps of editing ]] | [[Image:Edit_Operations.png|800px|thumb|center|Proposed steps of editing ]] | ||
| − | 1: '''Merge Small Gaps''' | + | |
| + | === 1: '''Merge Small Gaps''' === | ||
| + | |||
Goal: Create longest continuous trace segments by merging close endpoints | Goal: Create longest continuous trace segments by merging close endpoints | ||
Methods: Nearest neighbors (Closest end points), <br />Rejection based on conflicts and thresholds | Methods: Nearest neighbors (Closest end points), <br />Rejection based on conflicts and thresholds | ||
| Line 74: | Line 76: | ||
**Needs to be smoothed | **Needs to be smoothed | ||
| − | 2: '''Interpolate Large Gaps''' | + | |
| + | === 2: '''Interpolate Large Gaps''' === | ||
| + | |||
Goal: Connect Large gaps that step 1 cannot simply connect by addition of a single cell | Goal: Connect Large gaps that step 1 cannot simply connect by addition of a single cell | ||
Method: Larger gaps need new segments created, <br />new Trace Bits must be added, <br />smoothing operator. | Method: Larger gaps need new segments created, <br />new Trace Bits must be added, <br />smoothing operator. | ||
| Line 88: | Line 92: | ||
**Avoid creating edges | **Avoid creating edges | ||
| − | 3: '''Branch Points''' | + | |
| + | === 3: '''Branch Points''' === | ||
| + | |||
Goal: Detect and control Branching | Goal: Detect and control Branching | ||
Method: Detect most probable intercepts, <br />Determination of main branch and child, <br />Type of branch point | Method: Detect most probable intercepts, <br />Determination of main branch and child, <br />Type of branch point | ||
| Line 98: | Line 104: | ||
*Interpolation | *Interpolation | ||
| − | 4: '''Soma Detection''' | + | |
| + | === 4: '''Soma Detection''' === | ||
| + | |||
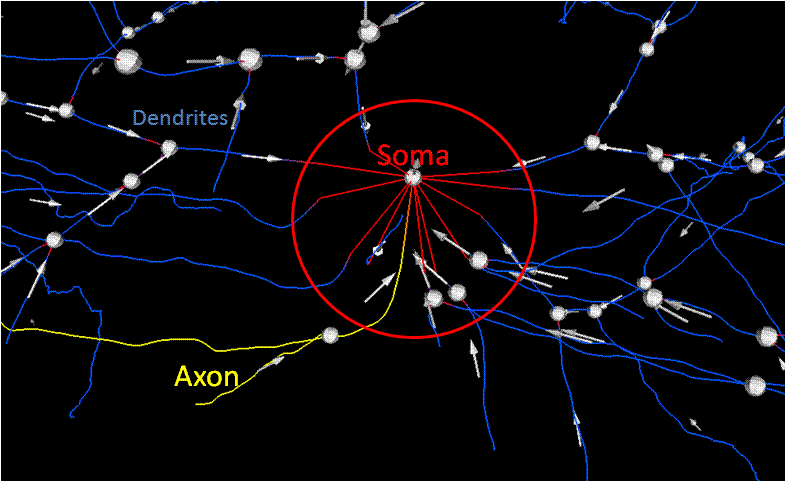
Goal: Correspond processes with a soma | Goal: Correspond processes with a soma | ||
Method: Segmentation of original data, <br />Localize the area to attach processes to soma, <br />Correct direction of traces | Method: Segmentation of original data, <br />Localize the area to attach processes to soma, <br />Correct direction of traces | ||
| Line 112: | Line 120: | ||
[[Image:FullConnections.gif|800px|thumb|center|Illustration of how the processes should be connected to a soma(shown in red). <br />The Dendrites are shown in blue, where the axon is shown in yellow.]] | [[Image:FullConnections.gif|800px|thumb|center|Illustration of how the processes should be connected to a soma(shown in red). <br />The Dendrites are shown in blue, where the axon is shown in yellow.]] | ||
| − | 5: '''Fragments''' | + | |
| + | === 5: '''Fragments''' === | ||
| + | |||
Goal: Removal of small traces that do not correspond to a process | Goal: Removal of small traces that do not correspond to a process | ||
Method: Small traces removed, <br />Leftovers from splitting operators, <br />Line fragments that cannot be merged | Method: Small traces removed, <br />Leftovers from splitting operators, <br />Line fragments that cannot be merged | ||
Revision as of 15:56, 3 July 2009
Contents |
Features and algorithms for Trace Editor
Feature List
| Feature | Description | Equation or Variable |
|---|---|---|
| Gap Size | Minimum distance between endpoints of two traces | 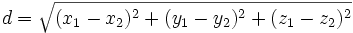
|
| Angle | The angle created between two traces normalized as vectors | 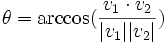
|
| Path Length | Total length along a trace, indicated by the size of the trace | 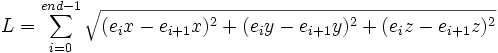
|
| Euclidean Distance | Straight line distance between the endpoints of a trace | 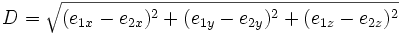
|
| Fragmentation Smoothness | Ratio of Path Length to Euclidean Distance[1] | 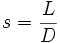
|
| Maximum Gap Distance | The maximum distance between endpoints that can be merged | Δ |
| Weights | The weights are used in the cost algorithm | α β |
| Cost | Weighted scalar evaluating the merge | 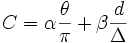
|
| Curvature | The Hessian matrix for checking branching point existence and interpolating the path. (Still under consideration) | ![\left[\begin{array}{ccc}
f(x)_{xx}(X) + \frac{\alpha}{2}f_{yy}(X) + \frac{\alpha}{2}f_{zz}(X) & (1-\alpha)f_{xy}(X) & (1-\alpha)f_{xz}(X) \\
(1-\alpha)f_{xy}(X) & f_{yy}(X) + \frac{\alpha}{2}f(x)_{xx}(X) +\frac{\alpha}{2}f_{zz}(X) & (1-\alpha)f_{yz}(X) \\
(1-\alpha)f_{xz}(X) & (1-\alpha)f_{yz}(X) & f_{zz}(X) + \frac{\alpha}{2}f(x)_{xx}(X) +\frac{\alpha}{2}f_{yy}(X)
\end{array} \right]](/mw/images/math/8/6/2/86265c277f698370dbb9b8bbf95c9619.png)
|
| Parent | Considering nerves branching from the soma are a tree like structure, a parent point can be used to find a cycle. |
Algorithms
The algorithms suggested are used to control the editing process allowing for rule based cluster editing. The Goal is to complete group editing in five steps.
1: Merge Small Gaps
Goal: Create longest continuous trace segments by merging close endpoints Methods: Nearest neighbors (Closest end points),
Rejection based on conflicts and thresholds
- Minimal distance between end points
- Angle between traces- The angle is measured between the two traces by computing the norm of their vectors and the dot product.
The equation is: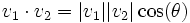 which when solved for θ is:
which when solved for θ is: 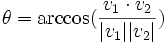
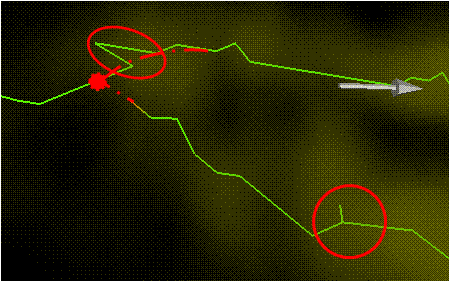
- Path length- The path length is considered if the end points are not the best fit for the merge. For instance, a merging that would cause a sharp peak in the neurite. This is a more serious problem for large gaps (see below). To help alleviate this for small gaps, merge at the point where the direction changes sharply.
- Gap distance is too large- The distance between the two points e_1, e_2 is computed to save computation. If the distance is longer than the excepted small gap distance, then the traces will not be considered for merging. A cost function adapted from "Automated Three-Dimensional Tracing of Neurons in Confocal and Brightfield Images" will take into account small gaps with two end points.
- Consider possibility of loops Though less probable in 3D, often there are 2D projections of 3D images resulting in loops being in the images. Loops in the branching structure can be avoided by checking the parents, as a trace cannot have two parents, but there can be two children.
- Angle between traces- The angle is measured between the two traces by computing the norm of their vectors and the dot product.
- Another trace is a better fit (Cost Function)
- Smallest gap
- Better Angular alignment
- "Bad merge"
- The merge causes corners
- Needs to be smoothed
2: Interpolate Large Gaps
Goal: Connect Large gaps that step 1 cannot simply connect by addition of a single cell Method: Larger gaps need new segments created,
new Trace Bits must be added,
smoothing operator.
- Curve fitting to find trend of:
- Direction
- Curvature
- Interpolation
- Extend the line
- Most probable vector
- Avoid creating edges
3: Branch Points
Goal: Detect and control Branching Method: Detect most probable intercepts,
Determination of main branch and child,
Type of branch point
- Distance maps
- Nearest neighbors (traces)
- Closest trace bits
- Angle of intercept
- Interpolation
4: Soma Detection
Goal: Correspond processes with a soma Method: Segmentation of original data,
Localize the area to attach processes to soma,
Correct direction of traces
- Image intensity
- Blob segmentation
- Centroid
- Distance map
- Connectivity
- Connected components
- Localization of processes
5: Fragments
Goal: Removal of small traces that do not correspond to a process Method: Small traces removed,
Leftovers from splitting operators,
Line fragments that cannot be merged
- Lowest percentile of length
- Traces with no parents or children
- Type dependent
References
Cite error:
<ref> tags exist, but no <references/> tag was found