Worm Analysis System
| Line 5: | Line 5: | ||
== System Overview== | == System Overview== | ||
| − | [[Image:Overview_new.png|center|799px| Figure 1: Main Window of the System]] | + | [[Image:Overview_new.png|thumb|center|799px| Figure 1: Main Window of the System]] |
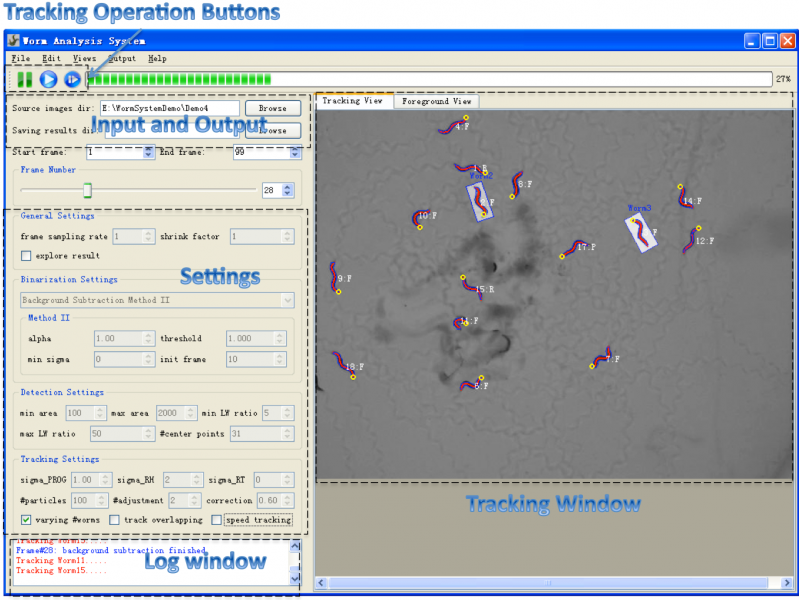
Figure1 shows a screenshot of the main window of the system. | Figure1 shows a screenshot of the main window of the system. | ||
*Menu: the menu provides operations like file operations and [http://farsight-toolkit.org/wiki/Worm_Analysis_System#Edit editing operations] which allow user to edit (reverse, delete and correct)selected worms in the tracking window. | *Menu: the menu provides operations like file operations and [http://farsight-toolkit.org/wiki/Worm_Analysis_System#Edit editing operations] which allow user to edit (reverse, delete and correct)selected worms in the tracking window. | ||
*Toolbar: the toolbr provides three buttons for controlling the tracking process. Users can pause tracking at any timepoint and | *Toolbar: the toolbr provides three buttons for controlling the tracking process. Users can pause tracking at any timepoint and | ||
do some observation or interactive editing when necessary. The thrid button make it possible for the user to start tracking from any saved tracking result which stores worm shapes, features and identities. Thus the user don't need to start over the tracking if the program was closed normally or abnormally. | do some observation or interactive editing when necessary. The thrid button make it possible for the user to start tracking from any saved tracking result which stores worm shapes, features and identities. Thus the user don't need to start over the tracking if the program was closed normally or abnormally. | ||
| − | [[Image:Toolbar.png|center|100px| Figure 2: Toolbar: pause tracking, start/resume tracking, start tracking from a saved point]] | + | [[Image:Toolbar.png|thumb|center|100px| Figure 2: Toolbar: pause tracking, start/resume tracking, start tracking from a saved point]] |
*Input and Output: the input and output section allows users to specify the directory of image sequence and the directory for storing tracking result. For each frame, the tracking result of it includes a txt file storing the worm structure(centerline and contour points), a excel file storing extracted features(motion state, shape and motion features, events, etc.) and a png image saved from the tracking window. The results could be explored at anytime during or after the tracking. | *Input and Output: the input and output section allows users to specify the directory of image sequence and the directory for storing tracking result. For each frame, the tracking result of it includes a txt file storing the worm structure(centerline and contour points), a excel file storing extracted features(motion state, shape and motion features, events, etc.) and a png image saved from the tracking window. The results could be explored at anytime during or after the tracking. | ||
*Settings: | *Settings: | ||
Revision as of 17:26, 16 February 2010
There is a compelling need for improved software systems for delineating the C."elegans" worms in time-lapse image sequences and tracking their movements. Segmentation and tracking provide the basis for making quantitative measurements of worm morphology, and various aspects of their dynamic behaviors. To facilitate the quantitative studies of worm behaviors, we developed a automated worm analysis system which is open-source and could be adopted by all researchers in the c."elegans" community.
Contents |
News
- 2010-02-14: Wiki Page for Worm Analysis System Created.
System Overview
Figure1 shows a screenshot of the main window of the system.
- Menu: the menu provides operations like file operations and editing operations which allow user to edit (reverse, delete and correct)selected worms in the tracking window.
- Toolbar: the toolbr provides three buttons for controlling the tracking process. Users can pause tracking at any timepoint and
do some observation or interactive editing when necessary. The thrid button make it possible for the user to start tracking from any saved tracking result which stores worm shapes, features and identities. Thus the user don't need to start over the tracking if the program was closed normally or abnormally.
- Input and Output: the input and output section allows users to specify the directory of image sequence and the directory for storing tracking result. For each frame, the tracking result of it includes a txt file storing the worm structure(centerline and contour points), a excel file storing extracted features(motion state, shape and motion features, events, etc.) and a png image saved from the tracking window. The results could be explored at anytime during or after the tracking.
- Settings:
- Tracking Window:
- Log Window
Overview
System Features
Multiple Linked Views
Edit
Settings
Keyboard Shortcuts
Explore Results
For information of our old worm project, please refer to [Old Worm Project].
References
- [1] Yu Wang, Roysam Badrinath, “Joint Tracking and Locomotion State Recognition of C.elegans from Time-Lapse Image Sequences,” IEEE International Symposium on Biomedical Imaging (ISBI) 2010 (oral presentation).
- [2] Roussel Nicolas, Morton Christine A, Finger Fern P, Roysam Badrinath, “A computational model for C. elegans locomotory behavior : Application to multiworm tracking,” IEEE transactions on biomedical engineering, pp. 1786-1797, 2007.
- [3] Katsunori Hoshi, Ryuzo Shingai, “Computer-driven automatic identification of locomotion states in Caenorhabditis elegans,” J Neurosci Methods, vol. 157(2), pp.355-363, 2006.
- [4] Huang KM, Cosman P, Schafer WR, “Machine vision based detection of omega bends and reversals in C. elegans,” Journal of Neuroscience Methods, vol. 158, Issue 2, pp. 323-336, 2006.
- [5] Nicolas Roussel, "A Computational Model for C.elegans locomotory behavior: Application to Multi-Worm tracking", Phd Thesis, 2007.
- [6] K.M. Huang, P.C. Cosman, and W. Schafer, “Automated tracking of multiple C. elegans with articulated models,” IEEE International Symposium on Biomedical Imaging (ISBI) 2007, pp. 1240-1243, 2007.
- [7] Fontaine, E., Barr, A., Burdick, J. W., “Model-based tracking of multiple worms and fish,” ICCV Workshop on Dynamical Vision, 2007.
- [8] Fontaine, E., Barr, A., Burdick, J. W., "Model-based tracking of multiple worms and fish", In ICCV Workshop on Dynamical Vision, 2007.
- [9] Wei Geng; Cosman, P.; Berry, C.C.; Zhaoyang Feng; Schafer, W.R., “Automatic tracking, feature extraction and classification of C. elegans phenotypes,” Biomedical Engineering, IEEE Transactions on, vol.51, no.10, pp.1811-1820, 2004.
- [10] Isard, M.; Blake, A., “A mixed-state condensation tracker with automatic model-switching,” Computer Vision, 1998. Sixth International Conference on, pp.107-112,1998.
- [11] G.J. Stephens et al., "Dimensionality and Dynamics in the Behavior of C. elegans", PLoS Comp. Biol., 2008.