|
|
| (159 intermediate revisions by 4 users not shown) |
| Line 1: |
Line 1: |
| − | [[Image:WTLView3.jpg|left|50px]] | + | '''For Our New Worm Analysis System, Please Refer to [[http://www.farsight-toolkit.org/wiki/Worm_Analysis_System Worm Analysis System]]''' |
| | <p align='justify'> | | <p align='justify'> |
| − | The Worm module of ''' FARSIGHT ''' represents a robust multiple worm segmenting and tracking locomotion events in live worm time – lapse sequences in the presence of a pheromone. Worms are tracked according to their distance from the pheromone spots and intrinsic features are computed from the image data for each worm. We present the application of a physically motivated approach to modeling the dynamic movements of the nematode C. elegans exposed to an external stimulus as observed in time-lapse microscopy image sequences. Specifically, we model deformation patterns of the central spinal axis of various phenotypes of these nematodes which have been exposed to pheromone produced by wild type C. elegans Worms are tracked according to their distance from the pheromone spots and intrinsic features are computed from the image data for each worm. The features measured include those proposed by Nicolas Roussel and those provided in the FARSIGHT Toolkit. </p> | + | |
| | + | |
| | + | ==Introductiom== |
| | + | The Worm module of ''' FARSIGHT ''' is a toolkit of computational methods designed to segment and track a population of C. elegans worms. The segmentation and tracking data can be used to identify locomotion events (e.g., pirouettes), and to quantify the impact of worm locomotion on external influences (e.g., pheromones). Worms are tracked according to their distance from the pheromone spots and morphological features are computed from the image data for each worm. We present the application of a physically motivated approach to modeling the dynamic movements of the nematode C. elegans exposed to an external stimulus as observed in time-lapse microscopy image sequences. Specifically, we model deformation patterns of the central spinal axis of various phenotypes of these nematodes which have been exposed to pheromone produced by wild type C. elegans Worms are tracked according to their distance from the pheromone spots and intrinsic features are computed from the image data for each worm. The features measured include those proposed by Nicolas Roussel and those provided in the FARSIGHT Toolkit. </p> |
| | | | |
| | <p align ='justify'>A model based algorithm for simultaneously segmenting and tracking of an entire imaging field containing multiple worms previously proposed by Nicolas Roussel has been implemented an applied to model worm behavior. An extension to the work propped by Roussel et. al., is to create a map of the worm’s behavior over time similar to the kymographs used by biologists to improve the accuracy of multiple hypothesis tracking by eliminating hypothesis based on a Euclidean distance metric to the worm kymograph. Interaction between worms is typically modeled as a random walk. We seek to give biologists a new tool to isolate the paths of peristaltic progression of C. elegans leads to unpredictable behaviors that are resolved using a variant of multiple-hypothesis tracking. The net result is an integrated method to understand and quantify worm interactions. | | <p align ='justify'>A model based algorithm for simultaneously segmenting and tracking of an entire imaging field containing multiple worms previously proposed by Nicolas Roussel has been implemented an applied to model worm behavior. An extension to the work propped by Roussel et. al., is to create a map of the worm’s behavior over time similar to the kymographs used by biologists to improve the accuracy of multiple hypothesis tracking by eliminating hypothesis based on a Euclidean distance metric to the worm kymograph. Interaction between worms is typically modeled as a random walk. We seek to give biologists a new tool to isolate the paths of peristaltic progression of C. elegans leads to unpredictable behaviors that are resolved using a variant of multiple-hypothesis tracking. The net result is an integrated method to understand and quantify worm interactions. |
| Line 8: |
Line 11: |
| | | | |
| | ==Modules== | | ==Modules== |
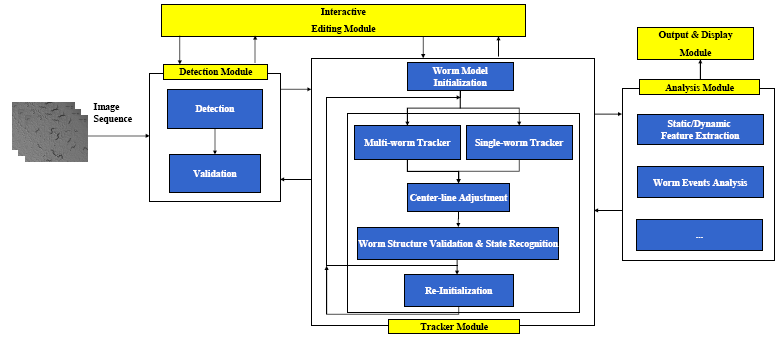
| − | [[Image:WormInteractionModel.jpg |left|800px]] | + | [[Image:Modules.png|thumb|center|800px| Figure 1: Modules for the Worm Tracking System]] |
| − | <br><br><br>
| + | |
| − | The modules of '''The Worm Project ''' are aimed at:
| + | |
| − | <br>
| + | |
| − | Exploiting complete worm track sequence to model movement and deformation in a correlated manner
| + | |
| − | <ul>
| + | |
| − | <li type = square>Modeling interaction and overlap using known locomotion events to exclude unfavorable hypothesis
| + | |
| − | <li type = square>Extending existing probabilistic approach to measurement and prediction using track summary</ul>
| + | |
| − | <br><br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | ===Detection===
| + | |
| − | Worms are modeled according to centerline and width profile.
| + | |
| − | Information about the entire image sequence is obtained by projecting the maximum intensity of
| + | |
| − | the sequence which is followed by a segmentation step.
| + | |
| − | <br><br>
| + | |
| − | Syntax:
| + | |
| − | WormBinarization [StringBeforeFilename] [MaskFilename] [StartFrame] [EndFrame][Levell]
| + | |
| − | TenickaSegment [StringBeforeInputFilename] [StringBeforeDataFilename] [LabeledImageFilename] [StartFrame] [EndFrame]
| + | |
| − | ====WormBinarization====
| + | |
| − | | + | |
| − | *WormBinarization accepts a series of grayscale images and returns a series of corresponding binary images
| + | |
| − | | + | |
| − | * The mask file used as an input is generated by performing a maximum intensity projection of the entire image sequence.
| + | |
| − | * The input images should be single-slice 8-bit TIFs. Other image formats have not been tested.
| + | |
| − | * The frame number must have n digits. We have tested this data with n == 4.
| + | |
| − | * The frame number must be at to the end of the image name.
| + | |
| − | | + | |
| − | '''Example Input'''
| + | |
| − | | + | |
| − | To track worms with the following image sequence...
| + | |
| − | C:\data\Im0000.tif
| + | |
| − | C:\data\Im0001.tif
| + | |
| − | C:\data\Im0002.tif
| + | |
| − | C:\data\Im0003.tif
| + | |
| − | C:\data\Im0004.tif
| + | |
| − | | + | |
| − | ...with their associated labeled image sequence...
| + | |
| − | C:\data\bw0000.tif
| + | |
| − | C:\data\bw0001.tif
| + | |
| − | C:\data\bw0002.tif
| + | |
| − | C:\data\bw0003.tif
| + | |
| − | C:\data\bw0004.tif
| + | |
| − | | + | |
| − | ...the following command should be used.
| + | |
| − | WormBinarization "C:\data\Im" "C:\data\bwIm" 0 4 0.145
| + | |
| − | | + | |
| − | '''Example Output'''
| + | |
| − | <br> <br>
| + | |
| − | [[Image:FarsightScreenshotForCelegans.jpg|center|thumbnail|450px| Screen Shot of the FARSIGHT Worm Module]]
| + | |
| − | <br>
| + | |
| − | <br>
| + | |
| − | | + | |
| − | | + | |
| − | | + | |
| − | See [[Features|FARSIGHT Features]]
| + | |
| − | <br><br>
| + | |
| − | | + | |
| − | ===Tracking===
| + | |
| − | Syntax:
| + | |
| − | Tracker [StringBeforeLabeledImageFilename] [StringBeforeRawImageFilename] [StartFrame] [EndFrame]
| + | |
| − | | + | |
| − | A center-line is extracted for all of the ‘tracks’ in the summary view of the image sequence.
| + | |
| − | <br>
| + | |
| − | The Euclidean distance from the worm center-line in each time step to the center-lines in extracted in
| + | |
| − | the summary view.
| + | |
| − | <br>
| + | |
| − | All hypothesis which are 1.2 times this distance are excluded from the possibilities used during tracking.
| + | |
| − | | + | |
| − | | + | |
| − | ===Features===
| + | |
| − | Worms are tracked according to their distance from the pheromone spots and intrinsic features are
| + | |
| − | computed from the image data for each worm.
| + | |
| − | <table class=MsoNormalTable border=1 cellspacing=0 cellpadding=0 width=646
| + | |
| − | style='width:484.2pt;border-collapse:collapse;border:none;mso-border-alt:solid black 1.5pt;
| + | |
| − | mso-yfti-tbllook:1184;mso-padding-alt:0in 0in 0in 0in;mso-border-insideh:.75pt solid black;
| + | |
| − | mso-border-insidev:.75pt solid black'>
| + | |
| − | <tr style='mso-yfti-irow:0;mso-yfti-firstrow:yes;height:30.05pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:1.5pt;border-left:
| + | |
| − | 1.5pt;border-bottom:1.0pt;border-right:1.0pt;border-color:black;border-style:
| + | |
| − | solid;mso-border-top-alt:1.5pt;mso-border-left-alt:1.5pt;mso-border-bottom-alt:
| + | |
| − | .75pt;mso-border-right-alt:.75pt;mso-border-color-alt:black;mso-border-style-alt:
| + | |
| − | solid;padding:.05in .1in .05in .1in;height:30.05pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'><b>Name</b></p>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'></p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:solid black 1.5pt;
| + | |
| − | border-left:none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-left-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-top-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:30.05pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'><b>Description</b></p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:solid black 1.5pt;
| + | |
| − | border-left:none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-left-alt:solid black .75pt;mso-border-top-alt:1.5pt;mso-border-left-alt:
| + | |
| − | .75pt;mso-border-bottom-alt:.75pt;mso-border-right-alt:1.5pt;mso-border-color-alt:
| + | |
| − | black;mso-border-style-alt:solid;padding:.05in .1in .05in .1in;height:30.05pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'><b>Formula</b></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Width</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Model of worm Width </p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\overline{w}\times(1-e^{-0.1\times(m-|2i-m|)})</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | | + | |
| − |
| + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Area</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Number of pixels in the worm</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>|\Omega|</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Minor Length</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Minor axis length of an ellipse with the same area as the worm which has been fit to our worm model</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>4\sqrt{\lambda_0}</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Major Length</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Major axis length of an ellipse with the same area as the worm which has been fit to our worm model</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>4\sqrt{\lambda_1}</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Eccentricity</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Ratio of maximum to minimum distance of the center of mass from the worm’s surface</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\cfrac{\max(\sqrt{||p-\overline{p}||})}{\min(\sqrt{||p-\overline{p}||})}</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Orientation</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Angle between the first principal axis of a given worm and the ‘x’ axis</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\arctan[\cfrac{dy}{dx}]</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>t_start</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Frame number that worm was initially detected</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>F_t(0)</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>t_end</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Frame number that worm was last detected</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>F_t(\infty)</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>t_length</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Total number of frames worm was tracked in a given image sequence</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\sum_t F_t, F = \# of frames</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Curvature</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>A metric to give an estimate of coiling as a worm behavior</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\kappa = (\cfrac{\theta_i}{L})</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Sum Squared Curvature</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Average Curvature squared, gives an estimate of the worm’s bending energy</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\sum_{i,t} mean(\cfrac{\theta_i^t}{L})</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Intensity</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>The average intensity of the pixels in a worm</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>mean(|\Omega|)</math></p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Peri_avg</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>The average worm speed</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>mean([u_i,...,u_n])</math> i=1,2,...,15
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| − | | + | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Peri_std</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>The standard deviation of the worm speed. Gives users insight into irregularities of worm peristaltic progression</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>std([u_i,...,u_n])</math> i=1,2,...,15
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| | | | |
| | + | ===The Worm Model=== |
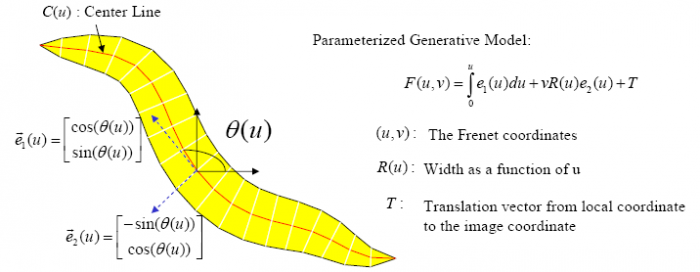
| | + | [[Image:WormModel.png|thumb|center|700px| Figure 2: Parameterized Generative Worm Model]] |
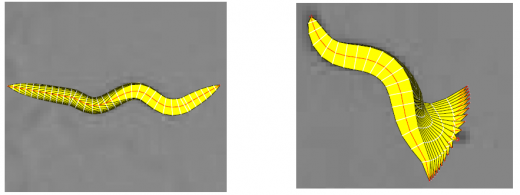
| | + | [[Image:Deformation.png|thumb|center|520px| Figure 3: Modeling the worm movement: Axial Progression and Radial Displacement]] |
| | | | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | In the tracking system, we combine and modify the two worm models used in [1][2] to make them more suitable for model-based multi-worm tracking, static/dynamic feature extraction and event analysis. This generative model could be deformed to model the two types of worm's crawling movement, as shown in Figure 3. |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>D_head_std</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Standard deviation of the displacement of the worm’s head</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>std(\gamma_k^H= {\tilde{\gamma}}^H \times e^{-0.09 \times (m-k)})</math>
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| | | | |
| | + | ===The Detection Module=== |
| | + | The tracker is automatically initialized by the detector. Worm detection generally involves:(1)Binarization and filling of small holes; (2)Extraction of the worm boundary based on the binary image; (3)Head and tail detection; (4)Center line and width computation; and (5)Instantiation of the worm model. In the detection module, detection is followed by the worm model validation to remove errors caused by spots in the background or overlapping worms. |
| | + | The detection module also corrects worms that were mis-tracked during the original tracking process. |
| | + | For more details about the detection module, please refer to: |
| | + | :*[http://farsight-toolkit.org/wiki/Detection_Module'''Detection Module''']: http://farsight-toolkit.org/wiki/Detection_Module |
| | | | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | ===The Tracking Module=== |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | The framework for the multi-worm tracker is shown in Figure 4. For more details about the tracker, please refer to: |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | :*[http://farsight-toolkit.org/wiki/Tracker_Module'''The Tracking Module''']: http://farsight-toolkit.org/wiki/Tracker_Module |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | [[Image:TrackerFramework.png|thumb|center|629px| Figure 4: Framework of the Tracker]] |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>D_tail_std</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Standard deviation of the displacement of the worm’s tail</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>std(\gamma_k^T= {\tilde{\gamma}}^T \times e^{-0.72 \times (k)})</math>
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| | | | |
| | + | ===The Analysis Module=== |
| | + | The tracking results can be fed to the analysis module for static/dynamic feature extraction, and event analysis. |
| | | | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | For a complete list of worm features, please refer to |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | :*[http://farsight-toolkit.org/wiki/Worm_Features%26Events'''Worm Features'''] |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Reversal Rate</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>The frequency of backward progression by the worm’s head</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\cfrac{||\sum u_i||}{\sum ||u_i||}</math>
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| | | | |
| | + | ===The Display and Editing Modules=== |
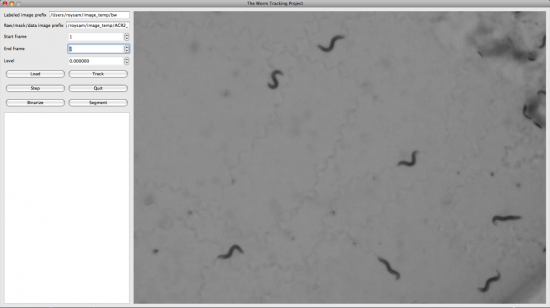
| | + | We also developed the display and editing modules for the Worm Tracker. The following screenshots were taken from them. |
| | + | [[Image:Mac-os-screenshot.png|center|thumbnail|550px| Screen shot of the tracker on Mac OS X]] |
| | + | [[Image:Before-tracking.png|center|thumbnail|550px| Screen shot of the tracker prior to starting to track]] |
| | + | [[Image:Seven-worms.png|center|thumbnail|550px| Screen shot with multiple worms selected]] |
| | | | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | ==Usage of the tracker== |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | For the console program for tracking without GUI, usage is as follows: |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | MWT.exe FileName Index1 Index2 GrayThreshold AreaThreshold1 AreaThreshold2 |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt; | + | BGsub OuputDir ImageFormat |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | FileName: Directory of the images |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Class</p>
| + | Index1: First index of the images |
| − | </td>
| + | Index2: Last index of the images |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | GrayThreshold: Threshold used for binarization |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | AreaThreshold1: Minimum object area size |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | AreaThreshold2: Maximum object area size |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | BGsub: 1 = true (compute background image first for images with cluttered background) |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Each Phenotype is given a separate class from 1-N</p>
| + | 0 = false (directly use thresholding and hole filling filter) |
| − | </td>
| + | OutputDir: Directory for storing the tracking result (a text file containing point lists, a labeled image, and a image |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | showing the tracking result) |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | ImageFormat: Format of the images |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>N/A</math>
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| | | | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | Example: MWT.exe E:/WormDemo/TestImages/LGC35ACR20 1 100 40 400 2000 1 E:/ bmp |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left: | + | MWT.exe E:/WormDemo/TestImages/LGC35ACR20 1 100 85 400 2000 0 E:/ bmp |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Bearing</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>The angle between the velocity vector and the spatial vector from the worm’s position to the center of the pheromone spot(chemical gradient)</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>\theta = \arctan[\cfrac{dv}{ds}]</math>
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| | | | |
| | + | '''Selection of parameters:''' |
| | + | Users need to choose several parameters related to the computation of binary images. The quality of the binary images is very important for the tracking system. |
| | + | * Threshold for binarization: Since the intensity of most worm images could be modeled as a mixture of N=2 Gaussian mixtures[1], when directly using the thresholding and hole filling filter for preprocessing, the threshold should be selected as the value separating the two Gaussian distributions, as shown in Figure 5. Intensities smaller than this threshold will be set to 1; the rest will be set to 0. When using a background image for subtraction, intensities larger than the selected threshold will be set to 1; this is the opposite of the behavior when the thresholding and hole filling filter is used directly. The threshold value also can be chosen based on the histogram. |
| | + | [[Image:Binarization1.png|thumb|center|650px| Figure 5: Use of an image histogram for selecting the threshold value]] |
| | + | * Minimum and maximum object area size: These are used to clean the background objects left after binarization. The user should try several values until desirable binary images are obtained. |
| | + | * Background subtraction: For images a with cluttered background, this should be set to 1 (true). A background image would be computed first; for images with a clean background, this can be set to 0 (false). In this case, no background image will be computed, and direct thresholding and hole filling will be used. |
| | | | |
| − | <tr style='mso-yfti-irow:1;height:35.85pt'>
| + | For some examples of choosing parameters to reach desirable binary image, please refer to: |
| − | <td width=178 valign=top style='width:1.85in;border-top:none;border-left:
| + | :*[http://farsight-toolkit.org/wiki/Choosing_Parameters'''Choosing Parameters''']: http://farsight-toolkit.org/wiki/Choosing_Parameters |
| − | solid black 1.5pt;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-alt:solid black .75pt;
| + | |
| − | mso-border-left-alt:solid black 1.5pt;padding:.05in .1in .05in .1in;
| + | |
| − | height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>Instantaneous Turning Rate</p>
| + | |
| − | </td>
| + | |
| − | <td width=347 valign=top style='width:260.45pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.0pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'>The change in orientation over time</p>
| + | |
| − | </td>
| + | |
| − | <td width=121 valign=top style='width:90.55pt;border-top:none;border-left:
| + | |
| − | none;border-bottom:solid black 1.0pt;border-right:solid black 1.5pt;
| + | |
| − | mso-border-top-alt:solid black .75pt;mso-border-left-alt:solid black .75pt;
| + | |
| − | mso-border-alt:solid black .75pt;mso-border-right-alt:solid black 1.5pt;
| + | |
| − | padding:.05in .1in .05in .1in;height:35.85pt'>
| + | |
| − | <p class=MsoNormal style='margin-bottom:6.0pt'> <math>[\cfrac{d\phi}{dt}]</math>
| + | |
| − | </p>
| + | |
| − | </td>
| + | |
| − | </tr>
| + | |
| | | | |
| | | | |
| | + | ===Python Implementation=== |
| | + | We have wrapped the above tracking system in a python script. A user can access the python script by opening a python window and importing the script called wormdemo.py. |
| | + | * From the Farsight GUI select View > Open Python Window to start the python interpreter. |
| | + | <gallery widths=400px heights=200px> |
| | + | Image:open_python.PNG |
| | + | Image:python.PNG |
| | + | </gallery> |
| | + | [[Image:FarsightWormDemo.jpg|thumb|center|800px| Figure 6: Python Script]] |
| | | | |
| − | </table>
| + | * Load the wormdemo python script |
| | + | * Place images in a directory within the current working directory |
| | + | current_path = os.getcwd() |
| | + | image_dir = "Images" |
| | + | os.chdir(current_path + os.sep + image_dir) |
| | + | * Note that the script takes the parameters from the modules listed above as arguments for the subprocesses in the script |
| | + | args = current_path + os.sep + "executable_fname " + fname_binary + " " + fname_base + " " + start_frame + " " + end_frame |
| | + | A user can change the arguments used in the script by setting the variables to reflect the image names set in the image directory |
| | + | * Choose an option from the list to perform |
| | + | * To view output, use the GUI shown above to load the xml result file generated by executing modules |
| | + | [[Image:Loadresult.jpg|thumb|center|600px| Figure 7: Using Farsight GUI to load results]] |
| | + | [[Image:FarsightOutputFromXml.JPG |thumb|center|900px]] |
| | | | |
| − | ====Behaviors==== | + | == References == |
| − | * Pierce-Shimomura JT, Morse TM, Lockery SR, '''"The Fundamental Role of Pirouettes in Caenorhabditis elegans Chemotaxis,"''' The Journal of Neuroscience. 1999 Nov; 19(21):9557–9569. | + | *[1] Nicolas Roussel, Badrinath Roysam, "A Computational Model for C.elegans Locomotory Behavior: Application to Multiworm tracking", IEEE Trans On Biomedical Engineering. Vol.54, No.10, Oct,2007. |
| | + | *[2] Fontaine, E., Barr, A., Burdick, J. W., "Model-based tracking of multiple worms and fish", In ICCV Workshop on Dynamical Vision, 2007. |
| | + | *[3] Fontaine, E., Lentink, D., Kranenbarg, S., Müller, U., van Leeuwen, J., Barr, A.H., Burdick, J. W. "Automated visual tracking for studying the ontogeny of zebrafish swimming", Journal of Experimental Biology, 211, 1305-1316, 2008. |
| | + | *[4] Nicolas Roussel, "A Computational Model for C.elegans locomotory behavior: Application to Multi-Worm tracking", Phd Thesis, 2007. |
is a toolkit of computational methods designed to segment and track a population of C. elegans worms. The segmentation and tracking data can be used to identify locomotion events (e.g., pirouettes), and to quantify the impact of worm locomotion on external influences (e.g., pheromones). Worms are tracked according to their distance from the pheromone spots and morphological features are computed from the image data for each worm. We present the application of a physically motivated approach to modeling the dynamic movements of the nematode C. elegans exposed to an external stimulus as observed in time-lapse microscopy image sequences. Specifically, we model deformation patterns of the central spinal axis of various phenotypes of these nematodes which have been exposed to pheromone produced by wild type C. elegans Worms are tracked according to their distance from the pheromone spots and intrinsic features are computed from the image data for each worm. The features measured include those proposed by Nicolas Roussel and those provided in the FARSIGHT Toolkit.
A model based algorithm for simultaneously segmenting and tracking of an entire imaging field containing multiple worms previously proposed by Nicolas Roussel has been implemented an applied to model worm behavior. An extension to the work propped by Roussel et. al., is to create a map of the worm’s behavior over time similar to the kymographs used by biologists to improve the accuracy of multiple hypothesis tracking by eliminating hypothesis based on a Euclidean distance metric to the worm kymograph. Interaction between worms is typically modeled as a random walk. We seek to give biologists a new tool to isolate the paths of peristaltic progression of C. elegans leads to unpredictable behaviors that are resolved using a variant of multiple-hypothesis tracking. The net result is an integrated method to understand and quantify worm interactions.
Experimental results indicate that the proposed algorithms allow for an integrated high throughput automated analysis of the locomotive behavior of C. elegans during exposure to a given pheromone. The results for this work are generated by a module of the FARSIGHT toolkit for segmentation and tracking of multiple biological systems. The features calculated during this work can be edited and validated using the tracking editor available in the FARSIGHT toolkit. Overall, the method provides the basis for a new range of quantification metrics for nematode social behaviors.
In the tracking system, we combine and modify the two worm models used in [1][2] to make them more suitable for model-based multi-worm tracking, static/dynamic feature extraction and event analysis. This generative model could be deformed to model the two types of worm's crawling movement, as shown in Figure 3.
The tracker is automatically initialized by the detector. Worm detection generally involves:(1)Binarization and filling of small holes; (2)Extraction of the worm boundary based on the binary image; (3)Head and tail detection; (4)Center line and width computation; and (5)Instantiation of the worm model. In the detection module, detection is followed by the worm model validation to remove errors caused by spots in the background or overlapping worms.
The detection module also corrects worms that were mis-tracked during the original tracking process.
For more details about the detection module, please refer to:
The framework for the multi-worm tracker is shown in Figure 4. For more details about the tracker, please refer to:
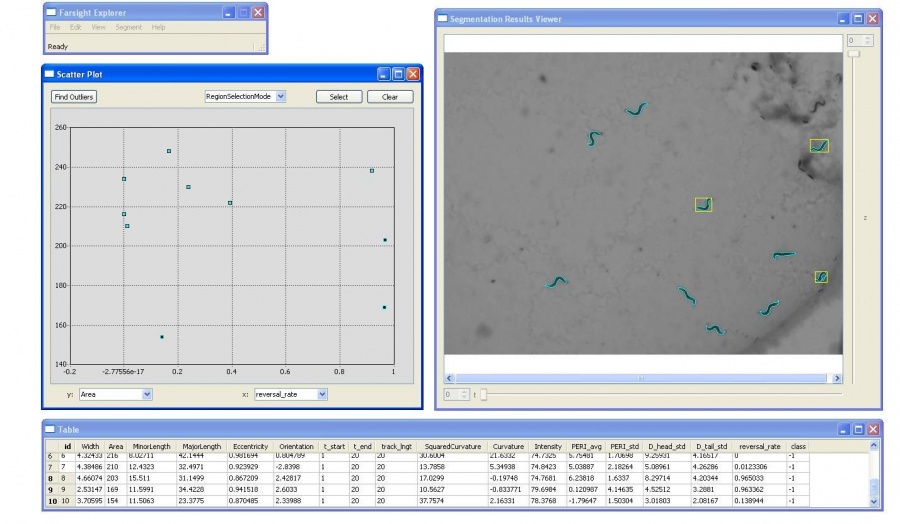
The tracking results can be fed to the analysis module for static/dynamic feature extraction, and event analysis.
We also developed the display and editing modules for the Worm Tracker. The following screenshots were taken from them.
For some examples of choosing parameters to reach desirable binary image, please refer to:
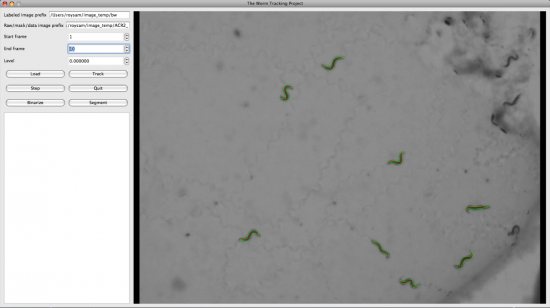
We have wrapped the above tracking system in a python script. A user can access the python script by opening a python window and importing the script called wormdemo.py.