The Worm Project
| Line 15: | Line 15: | ||
<br><br><br><br><br> | <br><br><br><br><br> | ||
====Detection==== | ====Detection==== | ||
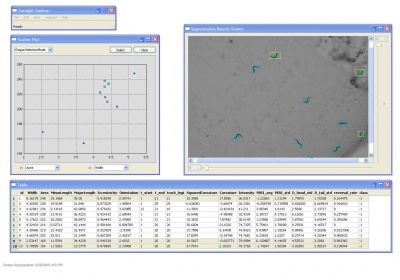
| + | [[Image:FarsightScreenshotForCelegans.jpg|right|400px| Screen Shot of the FARSIGHT Worm Module]] | ||
| + | <br> | ||
Worms are modeled according to centerline and width profile. | Worms are modeled according to centerline and width profile. | ||
Information about the entire image sequence is obtained by projecting the maximum intensity of | Information about the entire image sequence is obtained by projecting the maximum intensity of | ||
| Line 30: | Line 32: | ||
====Tracking==== | ====Tracking==== | ||
| − | |||
A centerline is extracted for all of the ‘tracks’ in the summary view of the image sequence | A centerline is extracted for all of the ‘tracks’ in the summary view of the image sequence | ||
The Euclidean distance from the worm centerline in each time step to the centerlines in extracted in | The Euclidean distance from the worm centerline in each time step to the centerlines in extracted in | ||
Revision as of 16:00, 25 April 2009
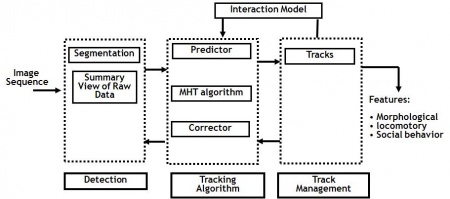
The Worm module of FARSIGHT represents a robust multiple worm segmenting and tracking locomotion events in live worm time – lapse sequences in the presence of a pheromone. Worms are tracked according to their distance from the pheromone spots and intrinsic features are computed from the image data for each worm. We present the application of a physically motivated approach to modeling the dynamic movements of the nematode C. elegans exposed to an external stimulus as observed in time-lapse microscopy image sequences. Specifically, we model deformation patterns of the central spinal axis of various phenotypes of these nematodes which have been exposed to pheromone produced by wild type C. elegans Worms are tracked according to their distance from the pheromone spots and intrinsic features are computed from the image data for each worm. The features measured include those proposed by Nicolas Roussel and those provided in the FARSIGHT Toolkit.
A model based algorithm for simultaneously segmenting and tracking of an entire imaging field containing multiple worms previously proposed by Nicolas Roussel has been implemented an applied to model worm behavior. An extension to the work propped by Roussel et. al., is to create a map of the worm’s behavior over time similar to the kymographs used by biologists to improve the accuracy of multiple hypothesis tracking by eliminating hypothesis based on a Euclidean distance metric to the worm kymograph. Interaction between worms is typically modeled as a random walk. We seek to give biologists a new tool to isolate the paths of peristaltic progression of C. elegans leads to unpredictable behaviors that are resolved using a variant of multiple-hypothesis tracking. The net result is an integrated method to understand and quantify worm interactions. Experimental results indicate that the proposed algorithms allow for an integrated high throughput automated analysis of the locomotive behavior of C. elegans during exposure to a given pheromone. The results for this work are generated by a module of the FARSIGHT toolkit for segmentation and tracking of multiple biological systems. The features calculated during this work can be edited and validated using the tracking editor available in the FARSIGHT toolkit. Overall, the method provides the basis for a new range of quantification metrics for nematode social behaviors.
Contents |
Modules
The modules of The Worm Project are aimed at:
- Exploiting complete worm track sequence to model movement and deformation in a correlated manner
- Modeling interaction and overlap using known locomotion events to exclude unfavorable hypothesis
- Extending existing probabilistic approach to measurement and prediction using track summary
Detection
Worms are modeled according to centerline and width profile.
Information about the entire image sequence is obtained by projecting the maximum intensity of
the sequence which is followed by a segmentation step.
Syntax:
WormBinarization [StringBeforeFilename] [MaskFilename] [StartFrame] [EndFrame][Levell] TenickaSegment [StringBeforeInputFilename] [StringBeforeDataFilename] [LabeledImageFilename] [StartFrame] [EndFrame]
See WormBinarization
See WormSegmentation
Tracking
A centerline is extracted for all of the ‘tracks’ in the summary view of the image sequence The Euclidean distance from the worm centerline in each time step to the centerlines in extracted in the summary view. All hypothesis which are 1.2 times this distance are excluded from the possibilities used during tracking.
Syntax:
Tracker [StringBeforeLabeledImageFilename] [StringBeforeRawImageFilename] [StartFrame] [EndFrame]
See Worm Tracker
Behaviors
- Pierce-Shimomura JT, Morse TM, Lockery SR, "The Fundamental Role of Pirouettes in Caenorhabditis elegans Chemotaxis," The Journal of Neuroscience. 1999 Nov; 19(21):9557–9569.