NucleusEditor
The main purpose of the NucleusEditor program is to view and segment images of nuclei, and then efficiently edit the segmentation using pattern analysis tools (PACE).
Contents |
Image Viewer
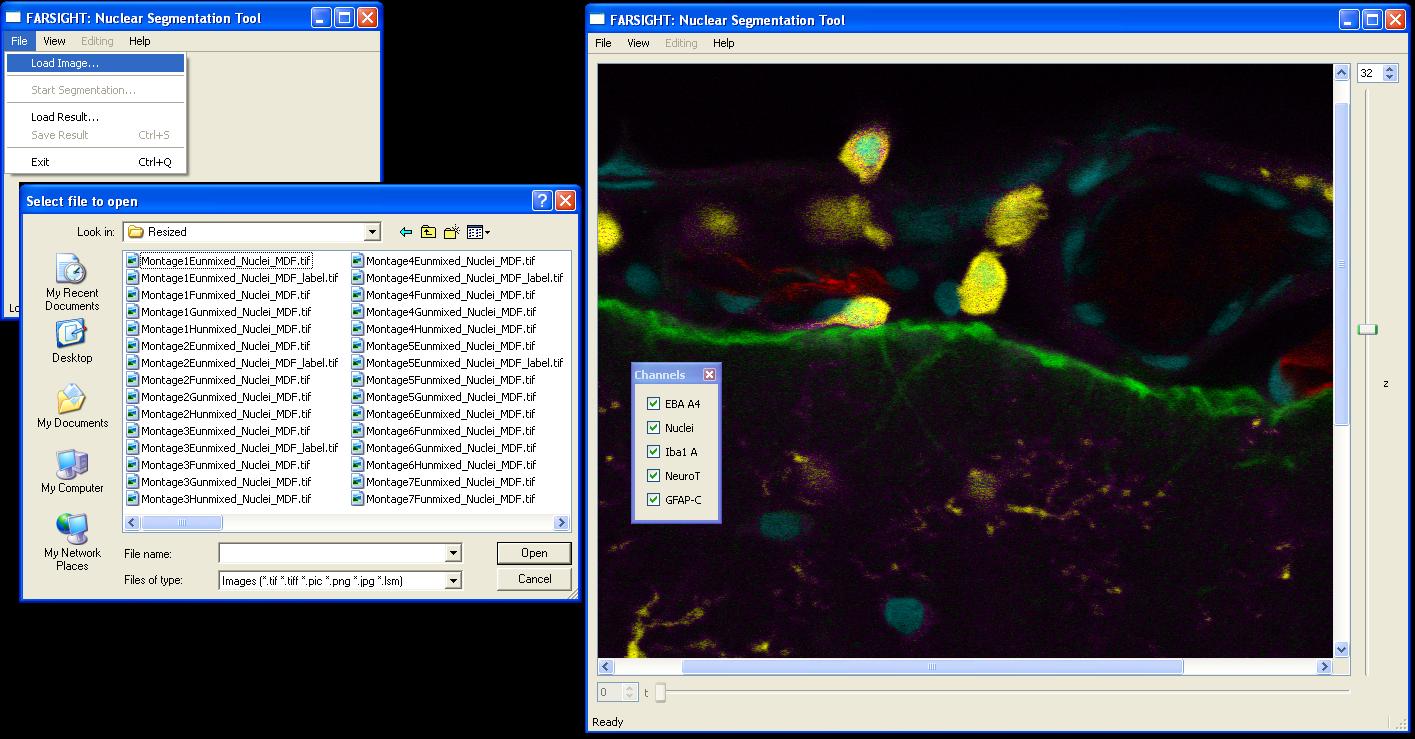
NucleusEditor allows users to view image data in a variety of file formats (tif, pic, lsm, etc). An image may be opened by selecting File|Load Image...
Segment Cell Nuclei
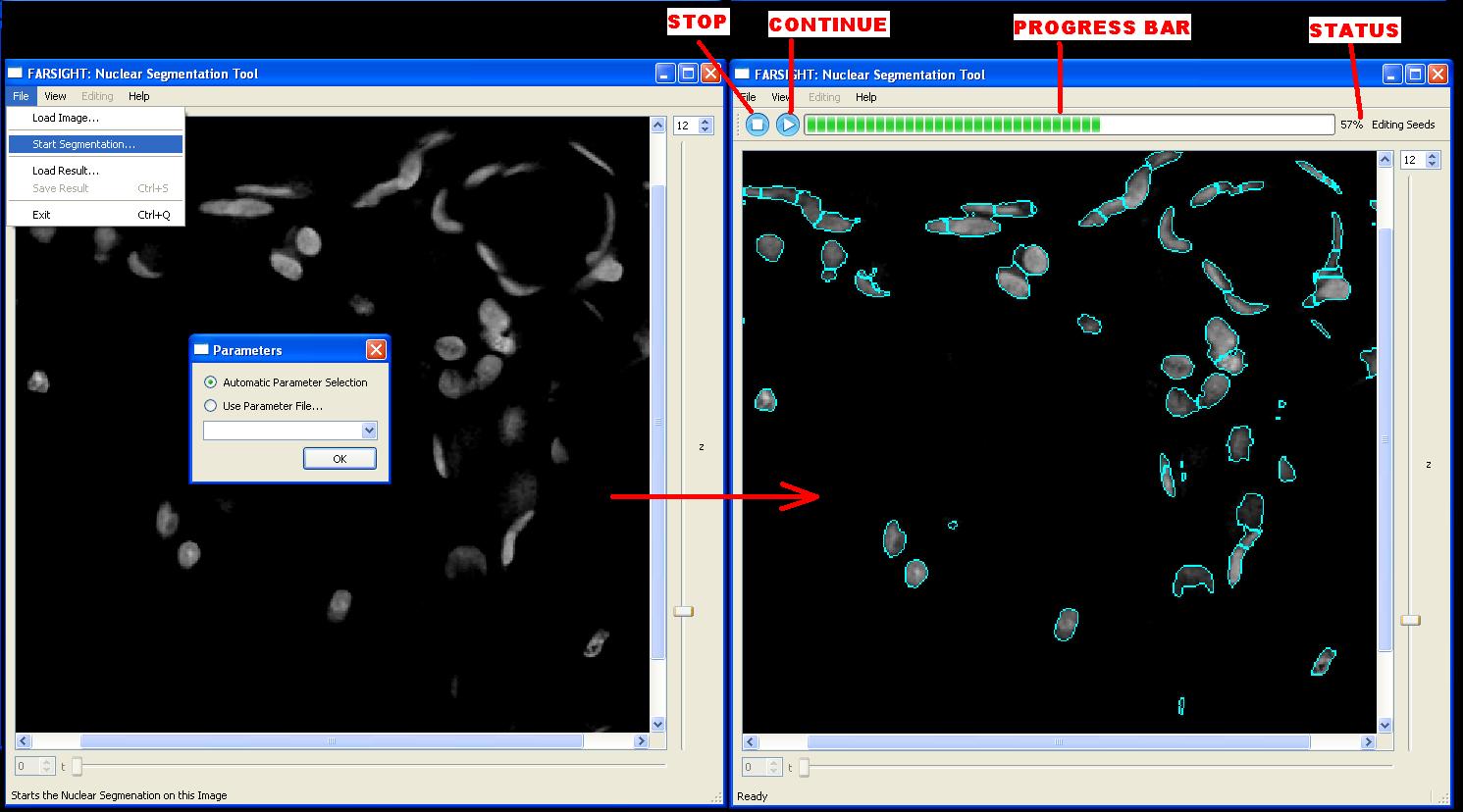
To begin segmentation, an image must already be loaded and visible in NucleusEditor. The nuclear segmentation algorithm is triggered by selecting File|Start Segmentation... The program will use the visible image for the segmentation algorithm. If the image has multiple channels, the program will search for the channel that contains the string "Nuclei" in its name. Before the segmentation algorithm can begin it must be initialized with a few parameters. These parameters can be chosen automatically using an innovative algorithm developed by Yousef Al-Kofahi, or they can be supplied in the form of a parameters file. The user must choose an option from the dialog box that will appear. When the segmentation begins, a new toolbar will be added to the NucleusEditor that shows the progress of the segmentation algorithm and provides a STOP button to cancel the segmentation.
Nuclear Segmentation is achieved in 3 major steps:
- 1. Initial Segmentation
- 2. Seed Editing
- 3. Final Segmentation
Upon completion of the initial segmentation, the program will pause and allow the user to inspect the result and edit the seeds [not available]. At this point the user may either continue segmentation (by pressing the "Continue" button), abort segmentation (by pressing the "Stop" button), or modify the parameters and start over (by changing the parameter file, pressing the "Stop" button, and then re-starting the segmentation using the saved parameter file).
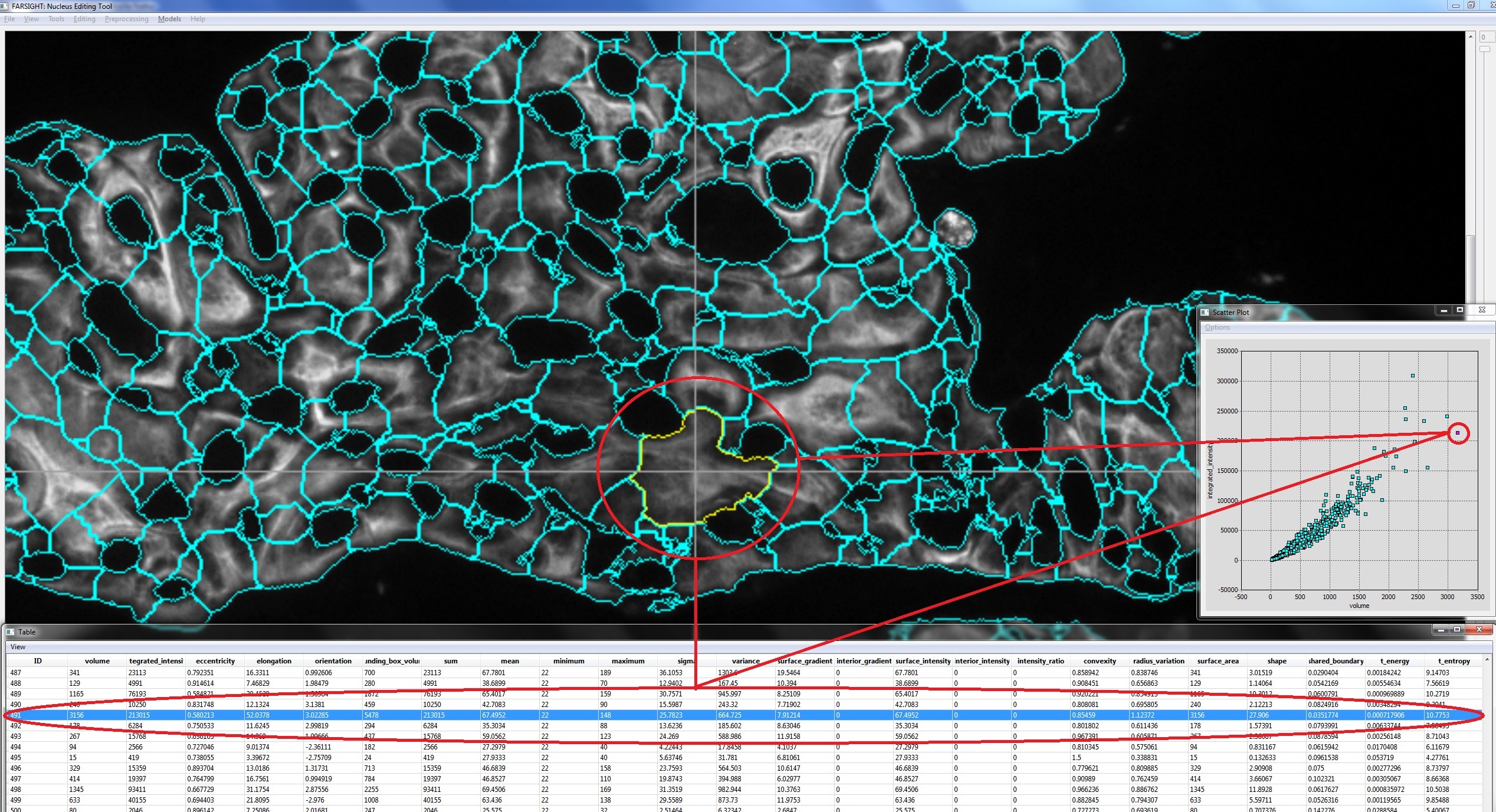
After the final segmentation, the program will compute features and show this metadata (mainly object features) using multiple data visualization tools that are all actively linked, allowing the data to be viewed in multiple spaces simultaneously. A cluster of objects to be selected based on any combination of operations in any of the multiple spaces. We term this method actively linked multiple spaces architecture (ALISA). NucleusEditor provides this linked-view architecture where objects can be visualized in image or geometric views, table views, plot views, and histogram views. Operations performed in one view are immediately visible in all of the views.
To save s segmentation result, select File|Save Result
In the ALISA mode a number of operations become active in each view:
- Segmentation Result
- Load nuclear segmentation result from file (File|Load Result...)
- Show/Hide Object Boundaries {View|Show Boundaries or Ctrl+B)
- Select/Deselect Objects {by clicking on them)
- Add Scatter Plot (View|New Scatter)
- Clear Selections (Editing|Clear Selections or Ctrl+C)
- Edit Functions (Edit Menu - see below)
- Scatter Plots
- Select/Deselect Objects
- Select/deselect by clicking on single object
- Clear all selections (Options|Clear Selections or Ctrl+C)
- Select group by holding CTRL key and creating a lasso via a series of clicks
- Options|Normalize - Ctrl+N (show features in normalized space)
- Tools|Find Outliers (ALISA - see below)
- Options (can change columns for X,Y and coloring)
- Select/Deselect Objects
- Table
- View|Sort table by... (To select column to sort by - in ascending order)
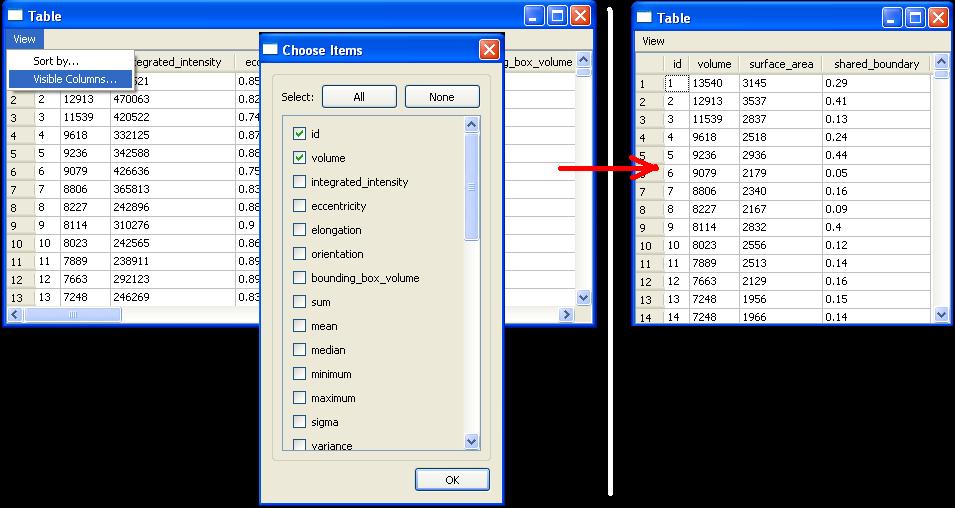
- Show/Hide table columns (View|Visible Columns)
Using the PACE Concept
Since automated segmentation algorithms are not perfect, there is a compelling need to develop efficient methods to identify and correct the automated segmentation errors. Due to the large size of the datasets, the identification and correction of the cells to be edited have to be done very fast. In order to achieve this, NucleusEditor implements kernel based pattern analysis algorithms where we can do group or cluster editing of multiple errors simultaneously. This dramatically reduces the amount of manual effort required compared to unassisted edit-based methods. We term this methodology PACE (Pattern Assisted Cluster Editing). By observing the nature of the errors, it will be possible to asses the performance of the segmentation algorithm used in NucleusEditor and modify it to reduce similar errors in the future.
ALISA is designed to allow group editing of objects. The idea is for the user to select a group of objects based on their features. This can be done manually, or using automated pattern analysis algorithms. Manual selection of objects is aided by scatter plots of features. The user can choose the features displayed along the x and y axes of scatter plots. To visualize more than 2 features at a time, ALISA allows the user to display an unlimited number of scatter plots. Each point on the scatter plot corresponds to a specific object in the image (with a unique identifier (ID)). Clicking the mouse on a point selects and highlights the corresponding object in all other views. The user can also draw a polygon on a scatter plot to select a group of objects visually.
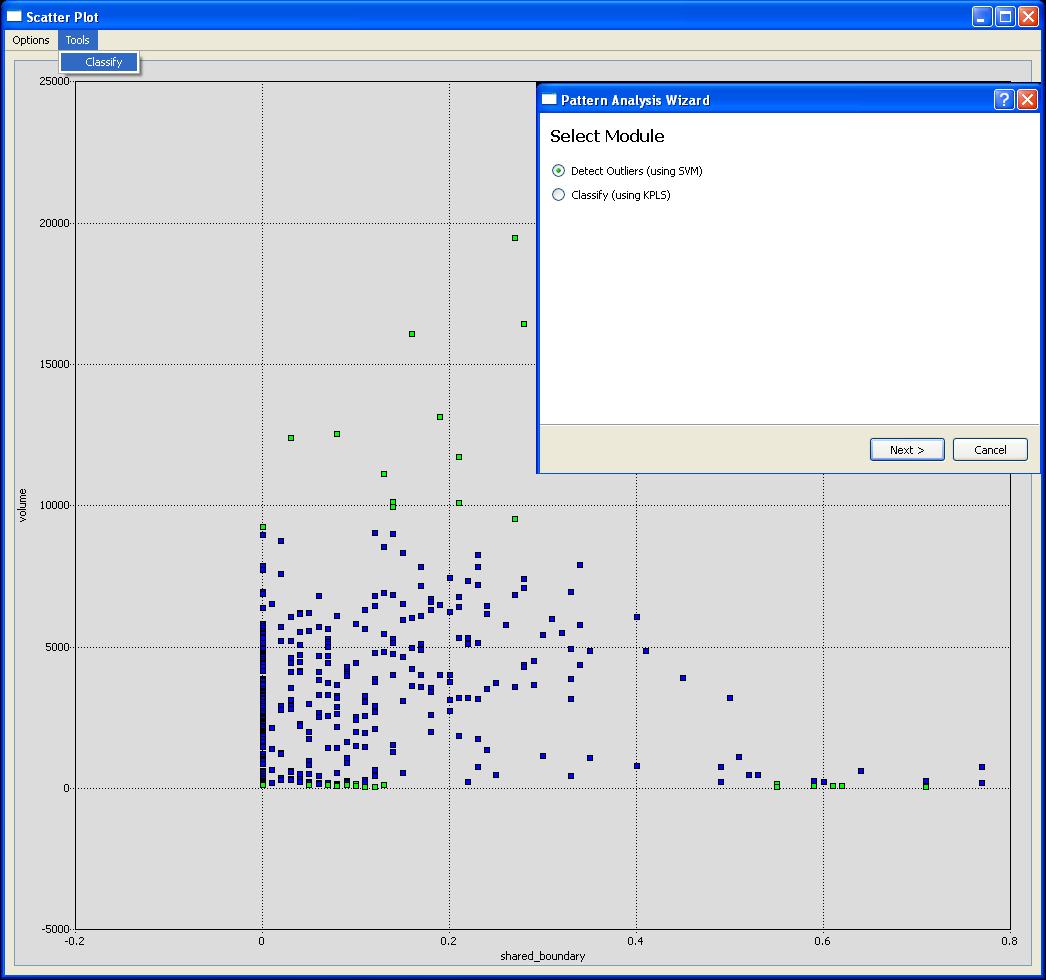
Computer-assisted selection of objects can also be performed. Features that highlight specific types of errors in segmentation (we call them diagnostic features) can also be defined and these objects selected and edited in their feature-space. Outlier detection tools are provided to assist in finding unusual objects to be inspected and/or edited. Outliers may correspond to segmentation errors (typical) or unusual cells (rare but possible). For outlier detection, the one-class Support Vector Machine (SVM) is employed [libSVM]. The one-class SVM proves to be a very useful tool for outlier detection, often highlighting a variety of quirky objects that should be inspected more closely, and edited when necessary. The outliers are shown in the segmentation, and scatter views, by a simple color coding scheme. The user can quickly locate, inspect and edit the outliers due to the linked nature of the views.
Clues
Two simple features have been shown to be particularly useful in detecting segmentation errors: volume and percent shared boundary. The following edit operations tend to correlate to these features in this way:
- Delete: small volume & zero (or small) percent shared boundary
- Merge: small volume & large percent shared boundary
- Split: high volume &| high surface area
Methods
Two main techniques can be used to locate objects that need to be edited: sorting by feature and outlier detection. In some cases the scatter plot showing outliers may be too crowded to select specific objects (ex. delete candidates), and it may be more useful to sort the table by a specific feature and select objects from this view.
- Sorting By Feature: The table view provides two very useful functions: View|Visible Columns and View|Sort By. The NucleusEditor provides a large wealth of feature data, most of which is noise to the editing process. Change the Visible Columns to only include those features that are useful while editing (ex. id, volume, shared boundary, surface area). Next we may want to find all of the objects with very small volume. On the scatter plot it is very hard to select just one of these objects and view it because they are tightly clustered together. By sorting the table by volume, it becomes very easy to find all of the objects with small volumes.
- Finding Outliers: The scatter plot view provides a tool for finding outliers in a set of data. This tool can be activated by selecting Tools|Find Outliers. The outlier tool uses the one-class SVM to find outliers. The "radius" of the outlier detector is modified by changing the Nu parameter. A larger Nu will result in a smaller radius (or more outliers). Please make sure that the Normalize box is checked. The outlier tool seems to be most effective when only 2 or 3 features are used. If the outliers are not immediately color-coded in the scatter plot it could be because the incorrect column for coloring is selected, please select the "outlier?" column by choosing Options|Set Color Column. From the scatter plot it is now easy to find sub-populations of objects that meet specific criteria (ex. small volume & high shared boundary - possible merge candidates).
Edits
At this stage in the process sub-populations of objects requiring specific edits should be identified, either by sorting the table or by finding regions (outliers) in the scatter plot. NucleusEditor provides the following three operations to edit cells, which will become active when a valid segmentation result is loaded:
- Delete
- Merge
- Split
The goal is to implement these operations using smart algorithms that allow them to be carried out in groups. These "smart algorithms" are still in development. The idea is to identify a large group of objects that need to be merged (for example), and simply tell the program to merge them. This could mean merging some of the selected objects with other selected objects, or it could mean merging a selected object with another, non-selected, object. Similarly, the split function should attempt to split an object into the most optimal number of objects with reasonable boundaries. As soon as objects are edited all views will be updated (however you will need to re-run the outlier detector to update outliers). Be sure to select File|Save Result to save results and any edits that have been performed.
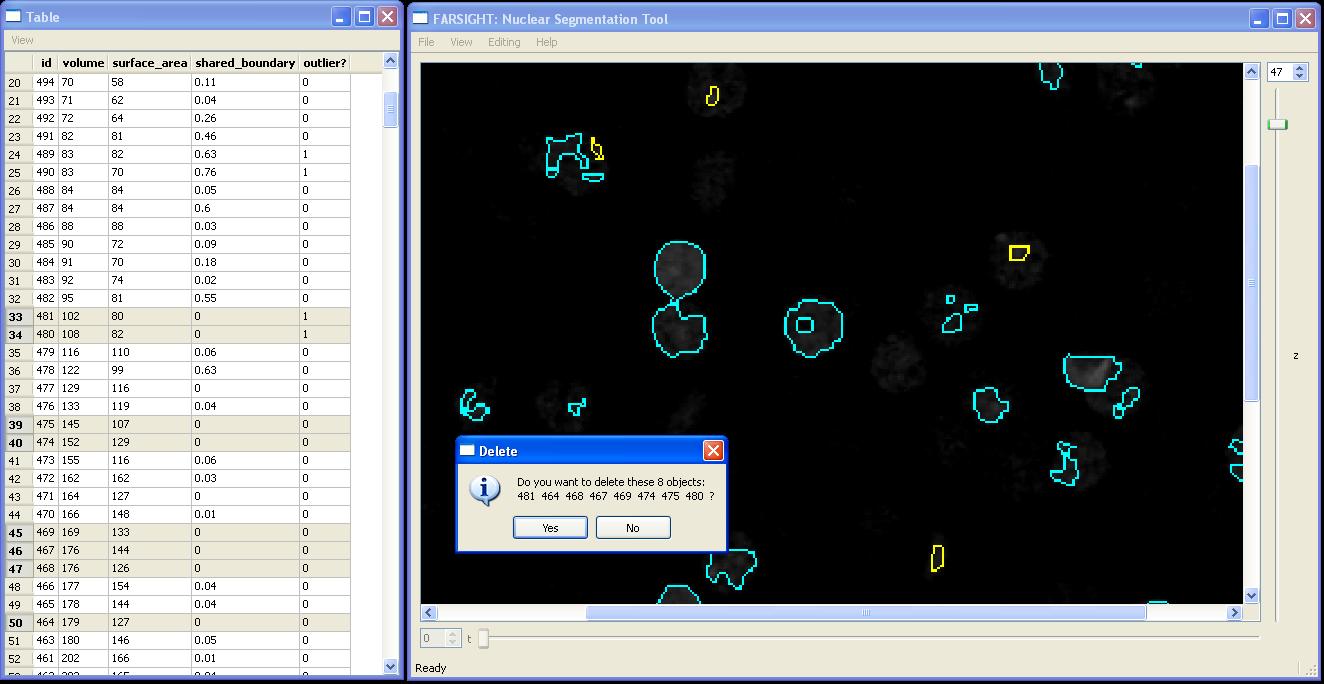
Delete
In order to delete objects we simply select them in any one of the available views and choose the "Editing|Delete Cells" menu item within the Nuclear Segmentation Window or press the "D" key. In the following example all cells with small volume and zero shared boundary will be deleted, they have first been selected in the table view.
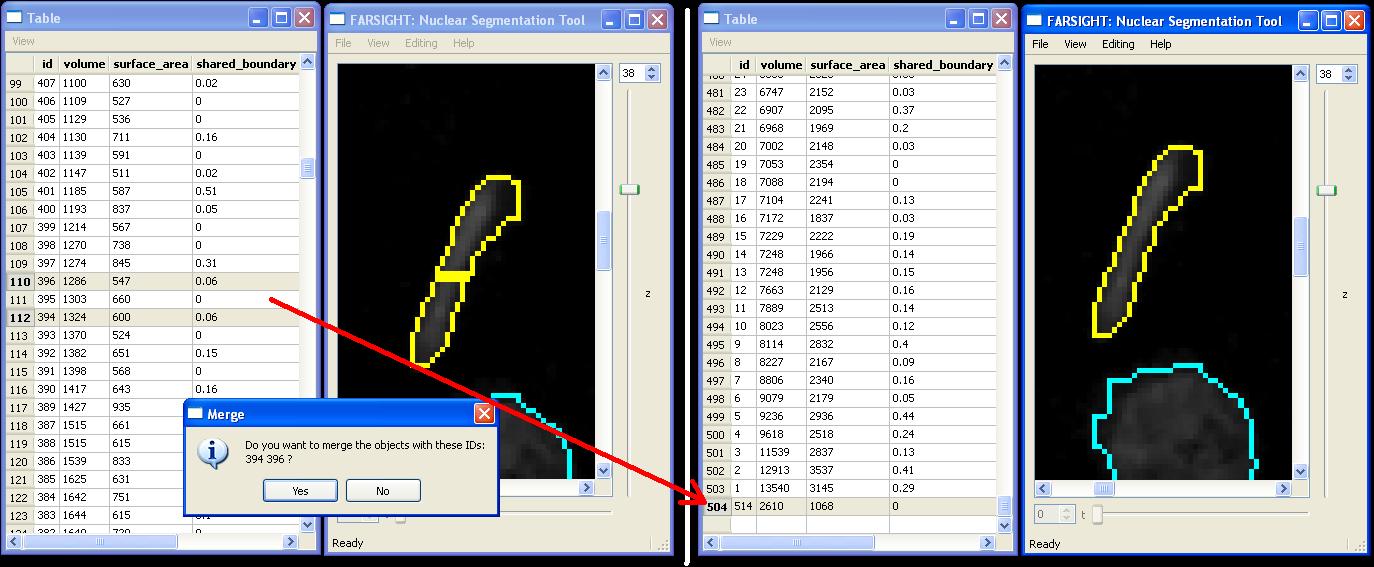
Merge
Merging objects requires the selection of two or more neighboring cells. Merging may be initiated from the "Editing|Merge Cells" menu item or by pressin the "M" key. Note that if the selected cells are not neighbors, they will not be merged. In this example we have merged object 394 and 396 to become a new object: 504.
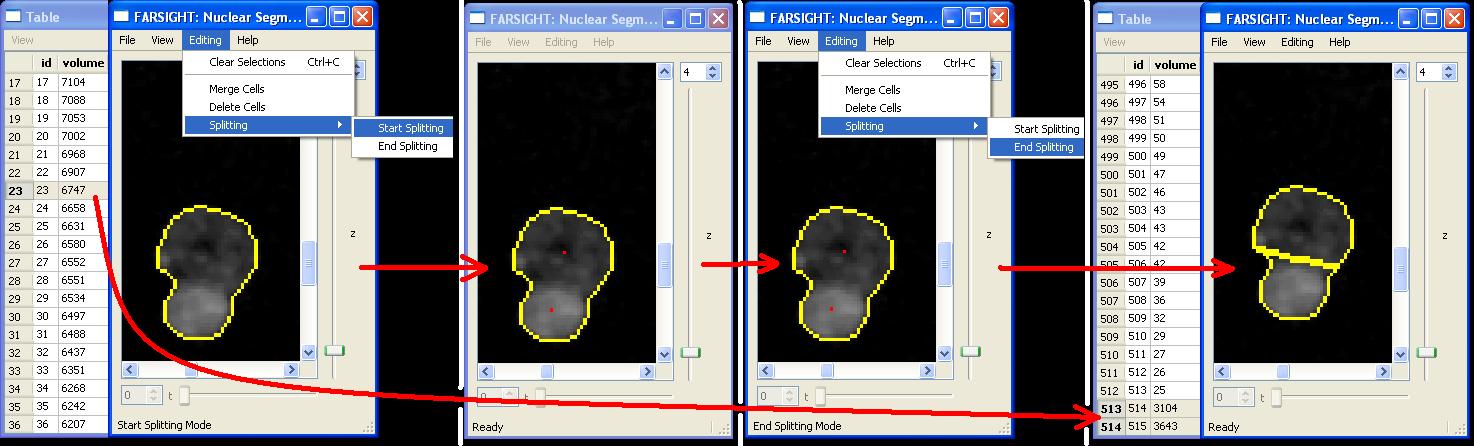
Split
Splitting objects is achieved by specifying two or more seed points. Start the splitting operation by selecting "Editing|Splitting|Start Splitting" from the menus. Left-click within the cell to place seed points (shown as red dots). Multiple objects may be split simultaneously by placing seed points in them. Once the seed points are specified, split the objects by choosing "Editing|Splitting|End Splitting" from the menu.Edit History Log
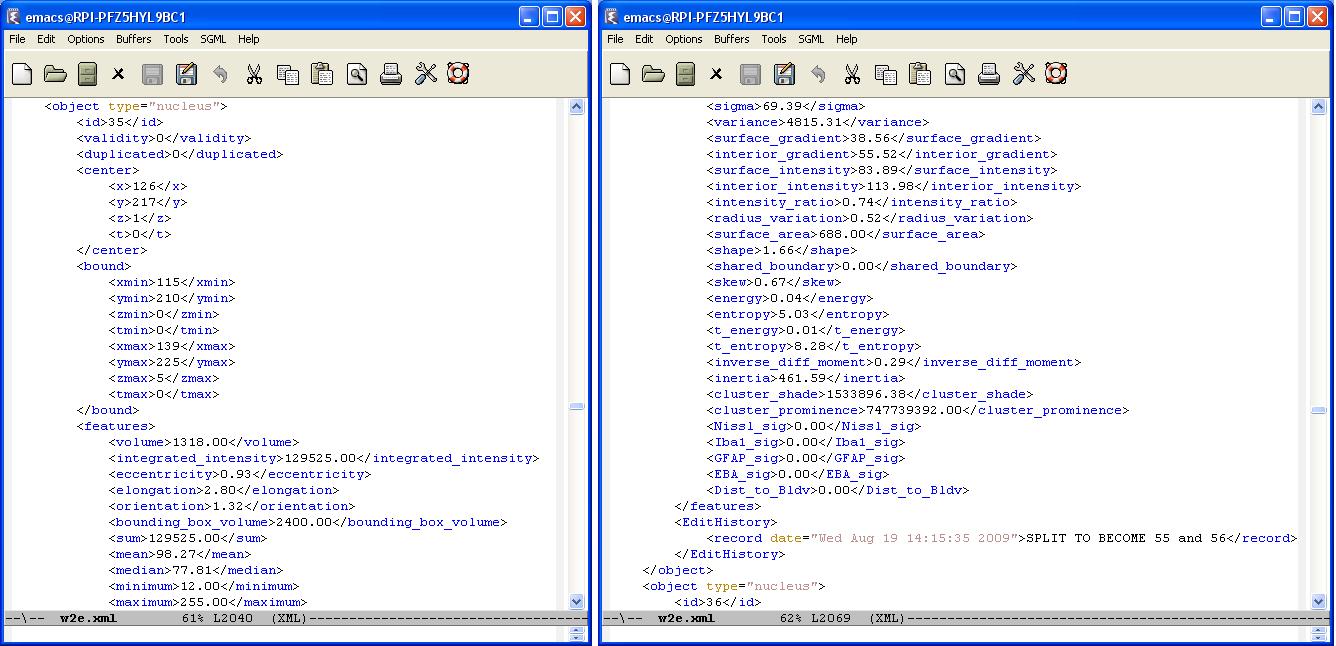
A typical XML result file consists of a set of objects whose structure is given in the following figure. Each object is defined with a set of features. In the following result file, an object corresponds to a cell nuclei which is identified by a unique id. A cell can be either valid or invalid depending on the editing operation. Initially all cells are valid and will be loaded. If a "delete" operation is applied to the cell , it will no longer be valid. But we still keep this cell data in the result file for logging purposes.
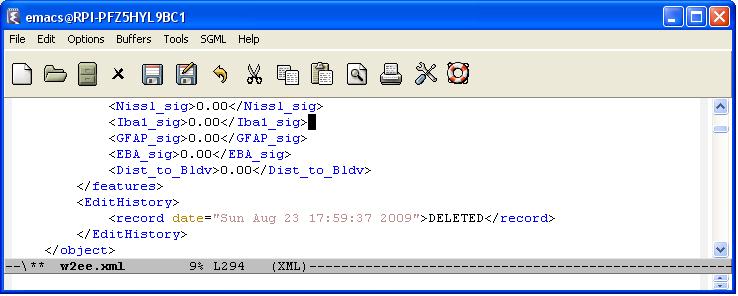
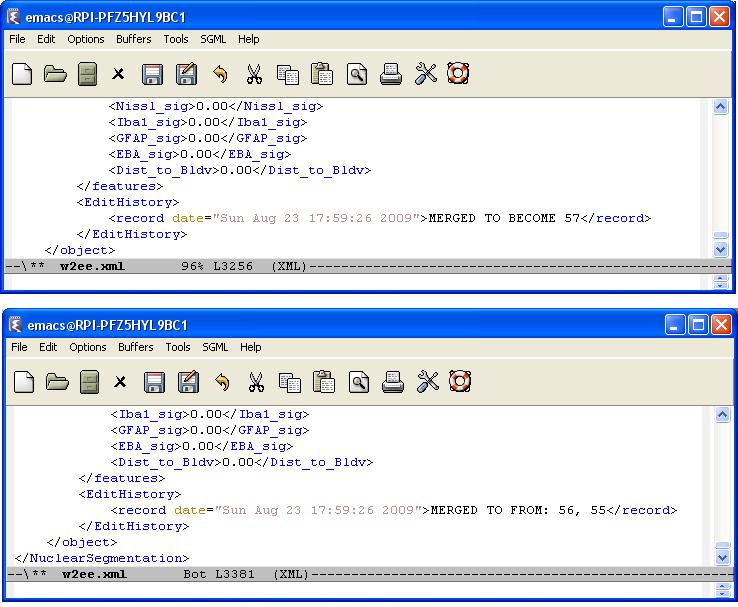
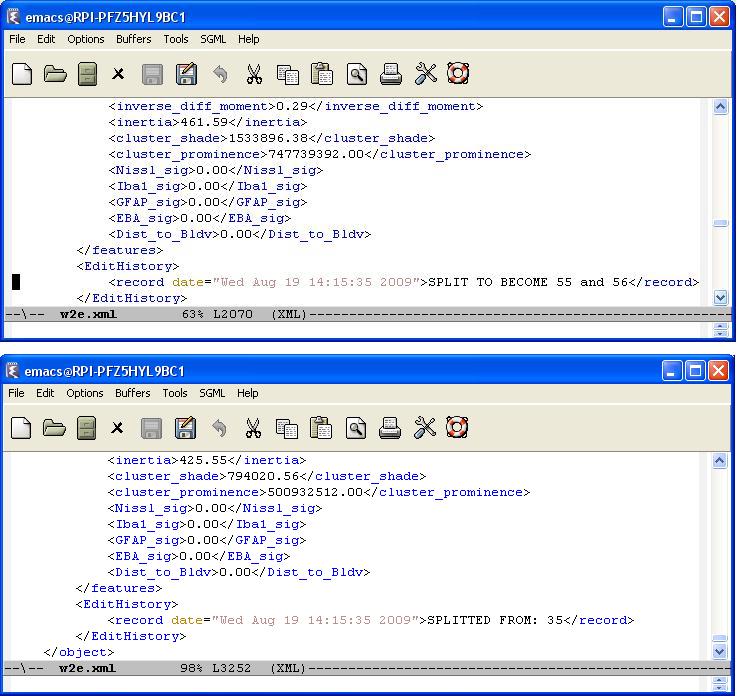
Every editing operation (delete, merge, or split) is recorded by creating an editing history log for each cell in the result file. We define an <EditHistory> element for each cell and list the editing operations with a proper timestamp and cell ids. There are three cases:
- Delete: We just note that this cell has been deleted (valid=0)
- Merge : In this case, two cells (id1, and id2) will be merged and a new cell will be created. Therefore, we first delete the first two cells and update their edit histories with this information, and then we update the edit history of the new cell noting that it is created by merging the cells with id1 and id2.
- Split : We note that two new cells will be created (with their ids) after splitting the cell of interest. In addition, the edit histories of each new cell will be updated with the id of the cell they are splitted from.
Summary
Once the corrected segmentation results are obtained with this way, hypothesis-driven and discovery driven research can be conducted by utilizing object features, associative object measurements, and the other tools that are part of the FARSIGHT_Toolkit.
This page was prepared by Isaac Abbott & Aytekin Vargun