Features
| (25 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | FARSIGHT contains a | + | FARSIGHT contains a broad and constantly growing library of software routines to compute diverse measurements from images. Measurements are also commonly referred to as "features" in the image analysis community. Broadly speaking, there are two classes of features in FARSIGHT intrinsic and associative. They are described below. |
'''1. Intrinsic Features: ''' Intrinsic features are measurements that quantify aspects of the morphology and/or appearance of objects in a single image channel. They can be computed based on a segmentation (object delineation) that results in a set of pixel/voxel labels. One must pay close attention to the units of the features. Also keep in mind the possibility of non-isotropic images - this is a common occurrence in biological microscopy. Usually, the axial dimension of a voxel is much greater than its lateral dimensions. To obtain feature measurements in physical units, one must scale pixel/voxel based feature values appropriately. The intrinsic measurements of each object are directly determined by its morphological class. Some examples are noted in the table below. | '''1. Intrinsic Features: ''' Intrinsic features are measurements that quantify aspects of the morphology and/or appearance of objects in a single image channel. They can be computed based on a segmentation (object delineation) that results in a set of pixel/voxel labels. One must pay close attention to the units of the features. Also keep in mind the possibility of non-isotropic images - this is a common occurrence in biological microscopy. Usually, the axial dimension of a voxel is much greater than its lateral dimensions. To obtain feature measurements in physical units, one must scale pixel/voxel based feature values appropriately. The intrinsic measurements of each object are directly determined by its morphological class. Some examples are noted in the table below. | ||
| − | {| class="wikitable" | + | {| class="wikitable"| border="1" |
|- | |- | ||
! Object Morphology | ! Object Morphology | ||
! Examples of Intrinsic Measurements | ! Examples of Intrinsic Measurements | ||
| + | ! Link | ||
|- | |- | ||
| Blobs | | Blobs | ||
| − | | location, diameter, volume, shape factor, surface area, eccentricity | + | | location, diameter, volume, shape factor, surface area, eccentricity, texture |
| + | | [[Intrinsic Features of Blobs]], [[3D_Subcellular_Location_Features|3D Subcellular Location Features]] | ||
|- | |- | ||
| Tubes | | Tubes | ||
| − | | centerline, surface locations, local diameter, branch points | + | | centerline, surface locations, local diameter, branch/crossover points, orientation |
| + | | | ||
|- | |- | ||
| Shells | | Shells | ||
| − | | thickness, shape factors | + | | thickness, shape factors, brightness |
| + | | [[Active Contours for Shell Segmentation]] | ||
|- | |- | ||
| Plate/Laminae | | Plate/Laminae | ||
| − | | thickness, surface area | + | | thickness, surface area, brightness |
| + | | | ||
|- | |- | ||
| Man-made objects | | Man-made objects | ||
| location, pose | | location, pose | ||
| + | | | ||
|- | |- | ||
| Foci | | Foci | ||
| location, brightness, diameter | | location, brightness, diameter | ||
| + | | | ||
|- | |- | ||
| Cloud | | Cloud | ||
| Brightness, texture | | Brightness, texture | ||
| + | | [[3D_Subcellular_Location_Features|3D Subcellular Location Features]] | ||
|- | |- | ||
|} | |} | ||
| − | + | The goal of FARSIGHT is to have a sufficiently rich, and well-documented library of intrinsic features available to the user for each morphological category. Another goal is to incorporate appropriate corrections when possible. For instance, most intensity/brightness measurements require a background correction step. Careful documentation of each feature, and clarifying its properties such as the units and invariance are an important goal. | |
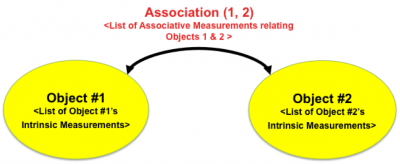
| − | '''2. Associative Features: ''' Associative features quantify associations/relationships between objects. They can be computed once objects have been delineated | + | '''2. Associative Features: ''' Associative features quantify associations/relationships between objects. They can be computed once objects have been delineated, and assigned unique identifiers (IDs). |
| − | + | [[Image:Association.png | center| 400px| Illustrating the notion of an associative measurement linking two objects. The list of associative measurements is dependent upon the nature of the two objects and the nature of the relationship between them that we are interested in quantifying. Graph theory offers a natural mathematical representation for associations. In this representation, intrinsic and associative become attributes of nodes and links, respectively.]] | |
| − | + | ||
| − | + | ||
| − | + | Some associative features involve objects and image data, details are [[Object_level_association|'''here''']]. For example, the spatial distribution of proteins around cell nuclei are quantified by sub-cellular location features[[http://www.farsight-toolkit.org/wiki/3D_Subcellular_Location_Features]]. Others relate objects to other objects, e.g., [[http://www.farsight-toolkit.org/wiki/TissueNets_Program TissueNets]]. The complex tissues of interest to us contain a dense web of interesting relationships, so we use a uniform graph-theoretic interpretation of associative measurements. An association can be described in terms of graphs in which each object is a node with a set of attributes (intrinsic measurements of the object), and each association is a link. Nodes and their attributes are uniquely identifiable by their object IDs. The attributes of each link is a list of measurements arising from associating the respective objects, and is uniquely identifiable by the ID’s of the associated objects. In principle, one can propose a large number of associations between a set of objects. However, a much smaller set of associations are biologically meaningful, plausible, relevant, and interesting. Once a graph-theoretic network of associations is defined, it can be queried and/or summarized in informative ways. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | The goal of FARSIGHT is to have a versatile set of 'programmable' computational methods available to the user. These are based on the simplest notions such as spatial proximity, neighborhood, and adjacency. The following paragraphs describe some of the kinds of associations we are interested in: | |
| − | + | ||
| − | [http:// | + | '''Raw Distance Based Associations: ''' The geometric distance between a pair of objects is a simple yet powerful basis for association. Until recently, large-scale distance computations were computationally expensive. Fortunately, developments in computational geometry have produced efficient algorithms for computing 3-D Euclidean distance maps. If the distance is zero (or close), the chosen image point is on/near the surface of the nuclei. A negative distance value indicates that the point is internal to the nuclear compartment, and a positive distance indicates that the point is outside. The location of the closest point on the nucleus also provides a readout of the object ID of the nearest nucleus, and thereby a link to all its properties. Distance maps help define a set of standard/custom zones associated with each compartment/ surface (inside/outside of a nucleus, small region outside a nucleus etc.). Zones are useful for efficiently computing zone integrals - the distance-weighted summation of a functional signal over a defined zone. Zone integrals are particularly valuable as a tool for classifying cells based on molecular markers. Visit [[http://www.farsight-toolkit.org/wiki/Object_level_association this page]] for more information on this topic. |
| + | |||
| + | '''Neighborhood Based Associations: ''' Spatial adjacency of neighboring structures is another powerful basis for defining associations among objects such as cells. At the next spatial scale, a number of biologically interesting questions involve the study of entire networks of cells. FARSIGHT includes the [[http://www.farsight-toolkit.org/wiki/TissueNets_Program TissueNets]] suite of tools to construct such networks 'on the fly' involving only the objects and relationships of interest. | ||
| + | |||
| + | '''Spatio-temporal Associations:''' Adjacency in time (across successive images of a time-lapse sequence) is an important form of association for dynamic studies. FARSIGHT incorporates tools for tracking objects over time to support this type of analysis. | ||
Latest revision as of 16:57, 17 March 2010
FARSIGHT contains a broad and constantly growing library of software routines to compute diverse measurements from images. Measurements are also commonly referred to as "features" in the image analysis community. Broadly speaking, there are two classes of features in FARSIGHT intrinsic and associative. They are described below.
1. Intrinsic Features: Intrinsic features are measurements that quantify aspects of the morphology and/or appearance of objects in a single image channel. They can be computed based on a segmentation (object delineation) that results in a set of pixel/voxel labels. One must pay close attention to the units of the features. Also keep in mind the possibility of non-isotropic images - this is a common occurrence in biological microscopy. Usually, the axial dimension of a voxel is much greater than its lateral dimensions. To obtain feature measurements in physical units, one must scale pixel/voxel based feature values appropriately. The intrinsic measurements of each object are directly determined by its morphological class. Some examples are noted in the table below.
| Object Morphology | Examples of Intrinsic Measurements | Link |
|---|---|---|
| Blobs | location, diameter, volume, shape factor, surface area, eccentricity, texture | Intrinsic Features of Blobs, 3D Subcellular Location Features |
| Tubes | centerline, surface locations, local diameter, branch/crossover points, orientation | |
| Shells | thickness, shape factors, brightness | Active Contours for Shell Segmentation |
| Plate/Laminae | thickness, surface area, brightness | |
| Man-made objects | location, pose | |
| Foci | location, brightness, diameter | |
| Cloud | Brightness, texture | 3D Subcellular Location Features |
The goal of FARSIGHT is to have a sufficiently rich, and well-documented library of intrinsic features available to the user for each morphological category. Another goal is to incorporate appropriate corrections when possible. For instance, most intensity/brightness measurements require a background correction step. Careful documentation of each feature, and clarifying its properties such as the units and invariance are an important goal.
2. Associative Features: Associative features quantify associations/relationships between objects. They can be computed once objects have been delineated, and assigned unique identifiers (IDs).
Some associative features involve objects and image data, details are here. For example, the spatial distribution of proteins around cell nuclei are quantified by sub-cellular location features[[1]]. Others relate objects to other objects, e.g., [TissueNets]. The complex tissues of interest to us contain a dense web of interesting relationships, so we use a uniform graph-theoretic interpretation of associative measurements. An association can be described in terms of graphs in which each object is a node with a set of attributes (intrinsic measurements of the object), and each association is a link. Nodes and their attributes are uniquely identifiable by their object IDs. The attributes of each link is a list of measurements arising from associating the respective objects, and is uniquely identifiable by the ID’s of the associated objects. In principle, one can propose a large number of associations between a set of objects. However, a much smaller set of associations are biologically meaningful, plausible, relevant, and interesting. Once a graph-theoretic network of associations is defined, it can be queried and/or summarized in informative ways.
The goal of FARSIGHT is to have a versatile set of 'programmable' computational methods available to the user. These are based on the simplest notions such as spatial proximity, neighborhood, and adjacency. The following paragraphs describe some of the kinds of associations we are interested in:
Raw Distance Based Associations: The geometric distance between a pair of objects is a simple yet powerful basis for association. Until recently, large-scale distance computations were computationally expensive. Fortunately, developments in computational geometry have produced efficient algorithms for computing 3-D Euclidean distance maps. If the distance is zero (or close), the chosen image point is on/near the surface of the nuclei. A negative distance value indicates that the point is internal to the nuclear compartment, and a positive distance indicates that the point is outside. The location of the closest point on the nucleus also provides a readout of the object ID of the nearest nucleus, and thereby a link to all its properties. Distance maps help define a set of standard/custom zones associated with each compartment/ surface (inside/outside of a nucleus, small region outside a nucleus etc.). Zones are useful for efficiently computing zone integrals - the distance-weighted summation of a functional signal over a defined zone. Zone integrals are particularly valuable as a tool for classifying cells based on molecular markers. Visit [this page] for more information on this topic.
Neighborhood Based Associations: Spatial adjacency of neighboring structures is another powerful basis for defining associations among objects such as cells. At the next spatial scale, a number of biologically interesting questions involve the study of entire networks of cells. FARSIGHT includes the [TissueNets] suite of tools to construct such networks 'on the fly' involving only the objects and relationships of interest.
Spatio-temporal Associations: Adjacency in time (across successive images of a time-lapse sequence) is an important form of association for dynamic studies. FARSIGHT incorporates tools for tracking objects over time to support this type of analysis.