Genetic Basis of Learning & Memory
The purpose of this page is to define our research goals and the demands that has on the functionality of the Farsight program. After speaking with Badri at our recent meeting, it is clear that there are two groups of changes that we as users would like to see implemented: there are changes that are required before we can apply Farsight to our research questions, and there are changes that are desired to make Farsight a more convenient tool in the workplace. The goal of this page is to communicate our needs so that the developers, who are divorced from the biology to an extent, can focus their efforts without having to waste time on features that are non-essential or could possibly be implemented at the researcher's end using scripting and so forth.
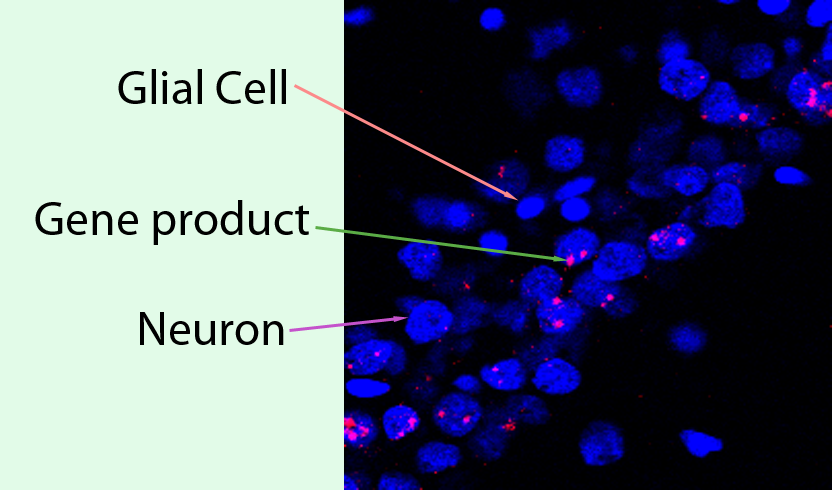
The Barnes lab is interested in the application of Farsight to biological samples using 2 or more channels of fluorescence. The tissue that we use is from rodent brain, and is typically processed with fluorescence in situ hybridization to visualize gene products that are expressed in behaviorally activated neurons. Our interest is to use Farsight to sort stained neurons into several classes depending on the presence of gene products. This requires proper segmentation of cellular nuclei and then the ability to look across multiple fluorescent channels to detect the presence of gene products in the spatial vicinity of cellular nuclei. This is an example of how our images often appear.
Within this image, there are two basic varieties of cell, the glial cell and the neuron. The nucleus of each cell is stained in blue. The second fluorescent channel, in red, is the mRNA produced for our gene of interest. We need to be able to extract the following information from such an image:
number of glial cell nuclei
number of neuronal nuclei
number of neurons expressing the gene's mRNA product
We first need to be able to separate all the cellular nuclei from each other. Then we need to be able to classify the cellular nuclei into "glial nuclei" and "neuronal nuclei". Finally, we need to be able to look at the second (and in some cases, third or fourth) channel and determine which nuclei are associated with gene product. The gene product may be inside the neuron's nucleus or outside the nucleus.
During Badri's visit, we determined a few changes that are required before we can use Farsight, as well as a few changes that are desired.
Required Changes
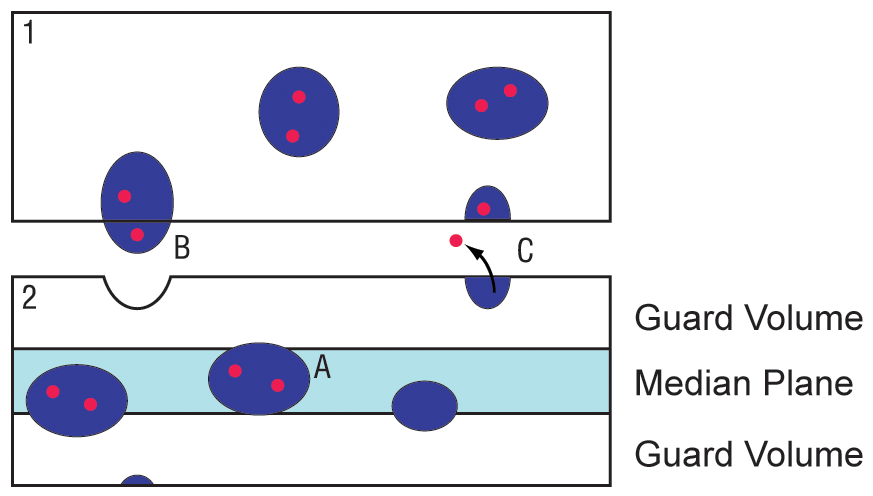
1) We need to be able to implement a proper median plane. In cell counting, the median plane means that we measure only that part of the tissue within the middle of the physical tissue slice. There are a few reasons that the edges of the tissue are not measured. The following figure illustrates why a median plane is important for biologists imaging cut tissue sections.
When a brain is cut into many very thin tissue sections, the edges cut by the knife are subject to a variety of changes that we refer to as cutting artifacts. Ideally, we want to measure the neurons that are expressing our gene product, as indicated by example A. When the neuron is situated in the middle of the cut tissue section, it is relatively preserved from physical damage by the knife.
Cases B and C illustrate a couple effects of knife cutting that make it difficult to detect a positive cell. In case B, the neuron is plucked by the knife into the top section (section marked 1) and is thus missing completely from the bottom section (marked 2). It is not practical in the lab to cut and stain every single section, so if only the bottom section is available for measurement, the positive cell that has been removed from the section will not be available to count and the measurements will be incorrect.
In Case C, the neuron is correctly cut in half by the knife, but unfortunately the contents of the cell have spilled out of the tissue section are forever lost. Once again if section 2 is the only section available to the researcher, this cell which should be counted as positive will not be included.
To avoid these sorts of errors in the quantification process, we implement what are called "guard volumes". For our purposes, a guard volume is implemented by defining the middle 20% of the tissue section- which should be preserved from damage by the knife- and count only the cells that are present in that 20%. In practice, this means identifying the optical sections within the confocal image stacks that are the top and bottom of the tissue section, and measuring only from the remaining middle 20% optical slices. The implementation of a median plane (not to 20% scale) is indicated in light blue in section 2. Only neurons that are sampled by the median plane are included in measurements- the cells that lie completely within the guard volumes are not counted. In this way we ensure that the cell counts are not affected by artifacts at the edge.
In Farsight, as Badri showed us at the recent meeting, a median plane can be implemented but only by setting an exclusion margin in the z plane that is symmetrical- that is, the user defines a margin of a set value, which is then implemented equally from the top and bottom of the stack. Unfortunately, during image collection, a variable number of slices may be collected above and below the actual physical slice. Therefore as users we need to be able to set the top and bottom of the tissue section within the optical stack manually, and then be able to apply a symmetrical exclusion margin using those planes instead of the limits of the optical stack itself.
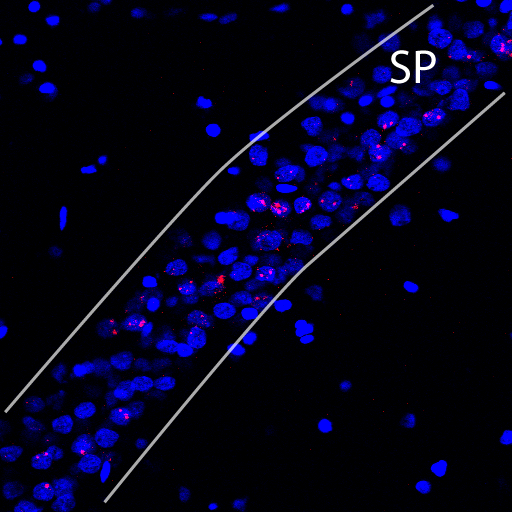
2) We need to be able to implement a region tool. For some confocal optical stacks, we may wish to quantify the entire x-y field of view. However, for other confocal stacks, there may be two or more different brain regions in the same image. As an example, in the image below, a confocal stack from a brain region called the hippocampus is depicted.
In the hippocampus, the neurons are arranged in a narrow layer called the stratum pyramidale (indicated by two white bounding lines and the abbreviation SP). We are absolutely not interested in the regions outside those bounding lines; in fact including those regions in any cell counts could bias our results. We need an implementation of a region tool that will allow us to manually exclude certain regions from a collected optical stack.
3) We will need to be able to associate features across channel data. We need to be able to not only segment the nuclei into neuronal nuclei and glial cell nuclei, but to also associate those segmented nuclei with the presence of fluorscence in a second (or third, etc.) channel.
Desired Changes
1) We need an improved segmentation of the nuclei. During Badri's demonstration, it appears that the segmentation is working pretty well. However in the case that two adjacent nuclei are included in one segmentation result, we need to be able to split the segmentation in a biologically accurate fashion. Currently it appears that splitting the segmentation works insofar that it takes one segmentation and produces two, but the resulting division does not follow a biological feature- it arbitrarily cuts the segmentation into two halves. If we were simply counting neuronal nuclei, this would be acceptable. Unfortunately we are interested in the association of gene product with specific cells; if the gene product lies in a part of the nucleus near the true border of the two cells, and the segmentation product after a splitting operation does not follow that true border, the gene product may be attributed to the wrong cell. I will include an image at a later date that better illustrates this issue.
2) It would be useful to be able to manually adjust the opacity of the boundaries marking the segmentation output, so that when we are evaluating the output for accuracy we can lower the opacity to better see the underlying pixels and cellular borders. Being able to adjust that opacity interactively would yield significant savings in time for the user as he/she judges the accuracy of the automated output.
3) It would be useful to be able to change the arbitrary color of the fluorescent channels within the Farsight program. In the images posted above, the nuclei are in blue and the gene products are in red; for some users, who may not be the ones that initially select the colors during image acquisition, it is difficult to discriminate the blue against the black background. Being able to select a different color for a channel would increase the usability of the software for investigators that are not collecting the images but instead are doing the quantification of the images. This division of labor is a common occurrence in the lab setting.
Note on Prioritization of Changes
These required and desired changes are all submitted with universal applicability in mind. That is, these are not specific only to the problems that our lab investigates. Any researcher that is sectioning brain tissue and wishing to use Farsight will need to be able to implement a median plane in their optical stacks; likewise, any researcher that is interactively assessing the automated output would benefit from being able to adjust the opacity of the overlaid boundaries. The separation of changes into "required" and "desired" categories is intended to assist in prioritizing the development of the software. As it stands with the version that Badri used to demo for us last month, we cannot use the software for our work until the required changes are implemented.