Morphologically Constrained Spectral Unmixing by Dictionary Learning for Multiplex Fluorescence Microscopy
Abstract
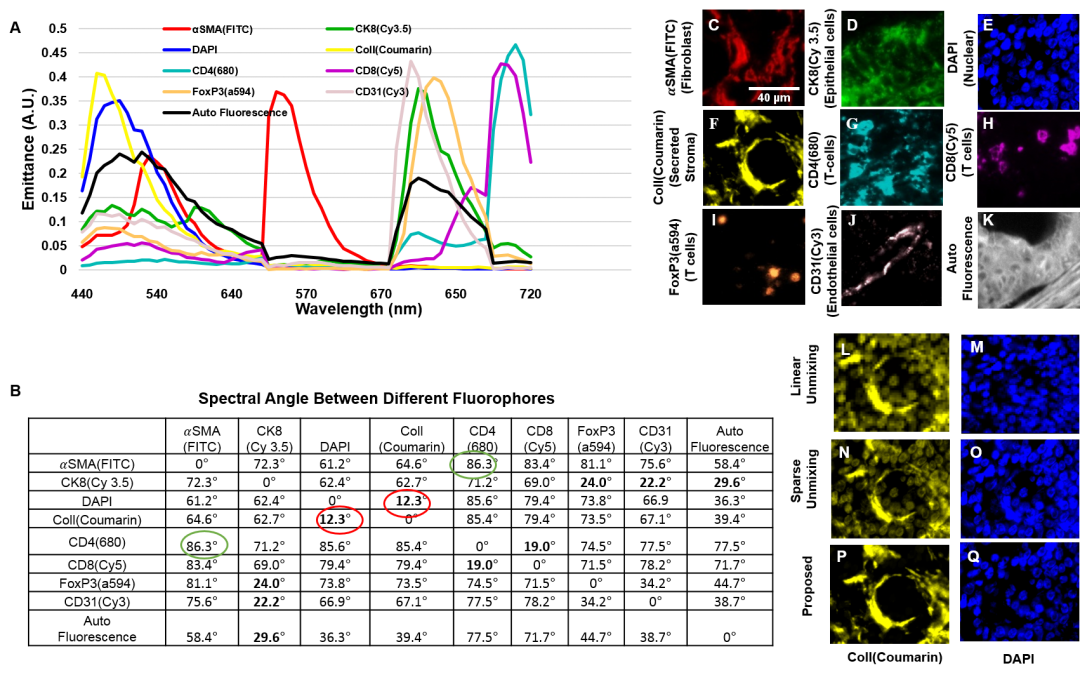
Motivation: Current spectral unmixing methods for multiplex fluorescence microscopy have a lim-ited ability to cope with high spectral overlap as they only analyze spectral information over individ-ual pixels. Here, we present adaptive Morphologically Constrained Spectral Unmixing (MCSU) al-gorithms that overcome this limitation by exploiting morphological differences between sub-cellular structures, and their local spatial context.
Results: The proposed method was effective at improving spectral unmixing performance by ex-ploiting: (i) a set of dictionary-based models for object morphologies learned from the image data; and (ii) models of spatial context learned from the image data using a total variation algorithm. The method was evaluated on multi-spectral images of multiplex-labeled pancreatic ductal adenocar-cinoma (PDAC) tissue samples. The former constraint ensures that neighbouring pixels corre-spond to morphologically similar structures, and the latter constraint ensures that neighbouring pixels have similar spectral signatures. The average Mean Squared Error (MSE) and Signal to Re-construction Error (SRE) ratio of the proposed method was 39.6% less and 8% more, respectively, compared to the best of all other algorithms that do not exploit these spatial constraints.
Availability: Specimen preparation and imaging protocols, open source software (MATLAB), and website (www.farsight-toolkit.org) Contact: B. Roysam (broysam@central.uh.edu)
https://github.com/megjhani/Unmixing_MCSU.git