Rationale for multi-dimensional microscopy
Many complex and dynamic tissue microenvironments play critical roles in health and disease – examples include stem-cell niches, brain tissue surrounding neuroprosthetic devices, retinal tissue, tumors, cancer stem-cell niches, and immune system components. Progress in these areas is much too slow compared to the need. Much of the biology remains unknown Therapeutic potential remains unrealized. Grand Challenges wait to be tackled. There is a compelling need to accelerate progress.
Why Bother with Imaging?
Imaging is the process of mapping an observable physical property over a region of space on a point-by-point basis. It is one of many investigational tools available, so it makes sense to have an accurate understanding of its 'value added'. As noted above, there is a need to speed up progress in the life sciences. Many high-throughput investigational technologies have been developed (and continue to be developed) to address this need. For example, many types of gene and protein microarrays have been developed that allow a large number of genes and proteins to be studied at once. This is powerful, but suffers from an important limitation. Obtaining this information requires one to 'grind up' all the cells. This process disrupts cellular structure, and loses all location information. We no longer know where a protein of interest is located within a cell. Obtaining spatial information requires imaging
Another high-throughput method is flow cytometry, that allows scientists to observe large numbers of cells in a short period. Tissues are broken up to the point that the cells of interest are suspended in a liquid medium. These suspended cells are then made to flow past optical sensors that record fluorescent signals. More advanced instruments even permit sorting of cells based on the recorded signals. Finally, some of the newer instruments, known as imaging flow cytometers can also record images of individual cells as they flow past the sensor. This method can preserve some amount of intra-cellular location information. However, flow-based methods inevitably disrupt tissue structure. We lose all information about the spatial arrangement and juxtaposition of cells in tissue. For many studies, this type of tissue context information is important.
In summary, methods that disrupt cell & tissue structure miss vital information on sub-cellular localization, location of cells in the tissue & relative to implanted devices (if any), structure of the cellular microenvironment, spatial relationships among cell & tissue entities, spatial dynamics of entities, and interactions. Only imaging of intact cells & tissue can provide these types of information.
Why Multi-dimensional Imaging?
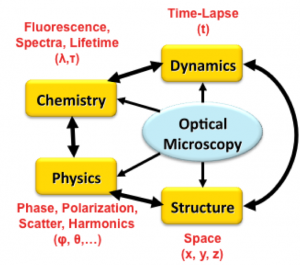
In a nutshell, multi-dimensional images offer dramatically more information about living systems, and offer important opportunities for advancing research. Let us examine some aspects of the information captured by multi-dimensional imaging. The figure below provides a graphical summary.
3-D Structure (Anatomy): Three-dimensional (x,y,z) (3-D) images of thick and intact tissue slices provide a far more accurate and complete description of the tissue anatomy compared to thin 2-D slices. Many aspects of cell function are intimately tied to their structure. The 3-D location of various entities within cells and tissue have an important influence on their function. Before the advent of 3-D microscopy, scientists had to infer 3-D structure from a series of 2-D slices, each of which conveys a very limited (and sometimes ambiguous) view of the 3-D structure of cells and tissue.
Chemistry: Molecular biology is all about understanding specific biochemicals in cells and tissue. Molecular imaging is the science of mapping the spatial distribution of specific molecular species (not individual molecules!) over a chosen region. Molecular imaging is accomplished using fluorescent labels that tag the structures of interest with a high degree of molecular specificity. Such tagging can also be made specific to the state of the molecule in question (e.g., phosphorylated or not). Fluorescence is a large and growing field with numerous specialties. For example, Fluorescent Resonance Energy Transfer (FRET) allows one to image molecules that are very close to each other. Fluorescence lifetime is a method that reveals the speed of the fluorescent response in addition to its spectral color.
Multiplexing: With rare exceptions, we are interested in the structure of more than one biological entity at a time. Cells live in a tissue environment composed of multiple cell types organized into multi-cellular units, that in turn have specific spatial relationships to other structures such as blood vessels. To make sense of these entities, we need the ability to see them together. Multiplexing is the idea of using two or more fluorescent labels of different spectral colors to tag multiple structures, and imaging them together to create a multi-color (multi-channel) image. Such four-dimensional imaging (x,y,z,λ) has the all-important advantage of preserving their spatial inter-relationships.
Dynamics: Living tissues are active. Many dynamic processes (cell migration, intra-cellular transport, etc.) in living cells/tissue can be imaged in the form of a time-lapse image sequence (movie). each "frame" of such a time-lapse movie can be 2-D or 3-D, or even 4-D. A time-lapse movie (image sequence (x,y,z,t)) reveals dynamic processes in the tissues. These dynamic processes occur in the context of the tissue habitat. By using additional channels to also record the tissue context using all of the available imaging dimensions (x,y,z,λ,t), we can observe living processes in their native tissue habitat. This is challenging - it requires ways to keep the cells alive on the microscope stage, ways to label the molecules of interest (usually done using fluorescent protein labeling), and ways to image with minimal phototoxicity (usually accomplished using multi-photon excitation).
Label-free Physical Properties: Many important physical properties of tissue can be accessed optically without the use of fluorescent (or such other extrinsic) labels. Examples include refractive index, polarization, and optical scatter (linear/non-linear).
Ongoing progress in this field is producing microscopes that can resolve much finer structures (even smaller than the Abbe limit), produce images much faster, and on a much larger scale. In the future, one can expect further growth in the number of possible dimensions. For instance, fluorescence lifetimes indicate molecular nano-environments, and the inclusion of additional modalities such as phase, polarization and non-linear scatter will undoubtedly provide additional data.