NucleusEditor
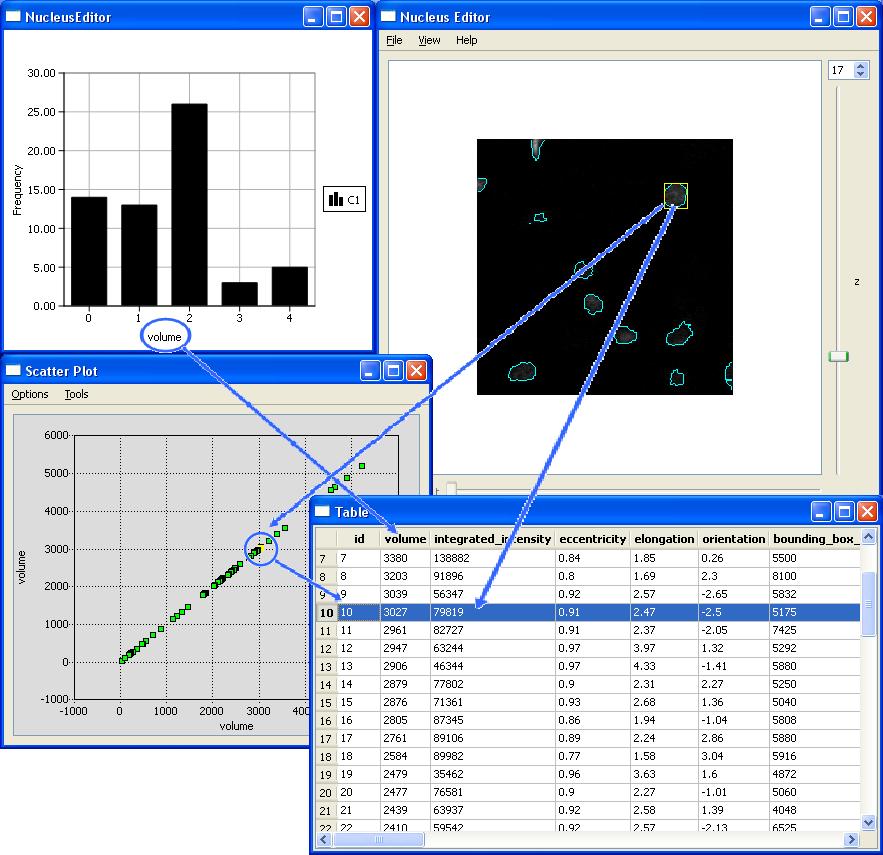
NucleusEditor allows users to view the image data, and the automatically generated multivariate metadata (mainly object features) using multiple data visualization tools that are all actively linked. This linkage is important, and allows the data to be viewed in multiple spaces simultaneously, and a cluster of objects to be selected based on any combination of operations in any of the multiple spaces. We term this method Actively linked multiple spaces architecture (ALISA). NucleusEditor provides this linked-view architecture where objects can be visualized in image or geometric views, table, views, plot views, and histogram views. Operations performed in one view are immidiately visible in all of the views.
In addition to viewing existing results, NucleusEditor makes it possible to segment images of of cell nuclei producing features with proper cell classifications.
Since automated segmentation algorithms are not perfect, there is a compelling need to develop efficient methods to identify and correct the automated segmentation errors. Due to the large size of the datasets, the identification and correction of the cells to be edited has to be done very fast. In order to achieve this NucleusEditor implements kernel based pattern analysis algorithms where we can do group or cluster editing of multiple errors simultaneously. This dramatically reduces the amount of manual effort required compared to unassisted edit-based methods. We term this methodology PACE (Pattern Assisted Cluster Editing). By observing the nature of the errors and analyzing them, it can be possible to asses the performance of the algorithms used in NucleusEditor for this purpose and modify them to prevent such errors in the future. We explain ALISA and Group Editing in more detail here.
This page was prepared by Aytekin Vargun (with Isaac Abbott's contributions)