Main Page
Audreys247 (Talk | contribs) (→RECENTLY ADDED PAGES) |
|||
| (152 intermediate revisions by 24 users not shown) | |||
| Line 1: | Line 1: | ||
| − | <big>'''Welcome to the | + | <big>'''Welcome to the FARSIGHT Project Wiki'''</big> |
| − | == | + | |
| − | * [[Features]] | + | <big>'''This Project is Developing Quantitative Tools for Studying Complex and Dynamic Biological Microenvironments from 4D/5D Microscopy Data'''</big> |
| − | * [[Registration page|Registration]] | + | |
| − | * [[TissueNets Program]] | + | The goal of the FARSIGHT project is to develop and disseminate a next-generation toolkit of image analysis methods to enable quantitative studies of complex & dynamic tissue microenvironments that are imaged by modern optical microscopes. Examples of such microenvironments include brain tissue, stem cell niches, developing embryonic tissue, immune system components, and tumors. Progress in mapping these microenvironments is much too slow compared to the need. Our knowledge of these systems has been painstakingly “pieced together” from large numbers of fixed, 2-D images of specimens revealing a small fraction of the molecular 'players' involved. The goal of this project is to help accelerate progress by: (i) harnessing the power of modern microscopy to help see the microenvironments in a much more detailed, direct, and comprehensive manner; and (ii) computational tools to analyze the multi-dimensional data produced by these microscopes. |
| + | |||
| + | '''Powered by this new Golden Age of optical microscopy: ''' Modern optical microscopes can capture multi-dimensional images of tissue microenvironments. First of all, these microscopes can record three-dimensional <math>(x, y, z)</math> images of thick, intact slices that are more realistic compared to thin slices. Next, they can record multiple structures simultaneously in a manner that preserves their spatial inter-relationships. This allows us to make '''associative measurements''' in addition to traditional morphological measurements (we call them '''intrinsic measurements'''). Such four-dimensional imaging <math>(x, y, z, \lambda)</math> is usually accomplished using multiple fluorescent labels that tag the structures of interest with a high degree of molecular specificity. Finally, it is now possible to capture such 3-D multi-channel images of living systems in the form of a time-lapse movie (image sequence <math>(x,y,z,t)</math>) that reveals dynamic processes in the tissues. Using all of the available imaging dimensions <math>(x, y, z, \lambda, t) </math>, we can now observe living processes in their native tissue habitat. Ongoing progress in this field is producing microscopes that can resolve much finer structures, produce images much faster, and on a much larger scale. In the future, one can expect further growth in the number of possible dimensions. For instance, fluorescence lifetimes indicate molecular nano-environments, and the inclusion of additional modalities such as phase, polarization and non-linear scatter will undoubtedly provide additional data. Click here to learn more about the [[rationale for multi-dimensional microscopy]]. To learn more about optical microscopy, [[http://micro.magnet.fsu.edu/primer/ Click Here]] | ||
| + | |||
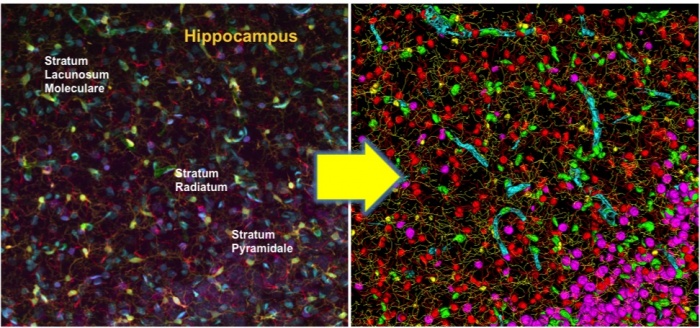
| + | [[Image:Hippocampus.jpg|center|thumb|700px| '''Cytovascular maps of brain tissue'''. One application of FARSIGHT is to map the structure of | ||
| + | complex tissues. Brain tissue is one of the most structurally complex. In this example, we use a multi-spectral confocal microscope, and innovative fluorescent labeling methods to collect three-dimensional 5-channel images of rat brain tissue. '''Color Code:''' ('''Cyan''') CyQuant-labeled cell nuclei; ('''Purple''') NeuroTrace-labeled Nissl substance; ('''yellow''') Iba1-labeled microglia; ('''red''') GFAP-labeled astrocytes; and ('''green''') EBA-labeled blood vessels. On the right is a frontal 3-D rendering of the combined multi-channel segmentation and cell classification results. Behind these visual results is a veritable cornucopia of '''intrinsic''' and '''associative measurements''' that can be queried to answer biological questions. To learn more, read our paper in the Journal of Neuroscience Methods [1] and/or browse this website. For a flowchart summary of the processing steps, [[Divide and Conquer Flow Chart|click here]].]] | ||
| + | |||
| + | '''It's a Toolkit, not a Software package: ''' We draw a distinction between these two words. A software package is a self-contained and tightly integrated software system that provides a defined set of services. A toolkit, on the other hand, is a collection of software '''modules''' with a set of standardized '''interfaces'''. To solve a given image analysis task, you can choose the right set of modules, and stitch them together using a scripting language ([[http://www.python.org Python]] in our case). Toolkits are easier to build and maintain (especially for academic laboratories like us), and more versatile since we cannot foresee all possible applications that FARSIGHT will encounter in the future. Click here to learn more about the [[FARSIGHT Toolkit]]. | ||
| + | |||
| + | '''The FARSIGHT Framework: ''' The [[FARSIGHT Framework]] serves as a conceptual guide to users and developers alike. The toolkit can be used to implement the framework by scripting modules together using the Python language. Click here to learn more about our [[long-term goals]]. | ||
| + | |||
| + | '''Open Source Software: ''' FARSIGHT is an open source toolkit, and you can view our works in progress. You can also contribute to this project. FARSIGHT draws upon major open source toolkits especially the [[http://www.itk.org/ Insight Toolkit (ITK)]], the [[http://www.vtk.org/ Visualization Toolkit (VTK)]], [[http://www.openmicroscopy.org Open Microscopy Environment (OME]]), and various others. The FARSIGHT community is composed of users in the life sciences and developers in the computational sciences (happily, many colleagues seem at ease in both categories!). We hope that our developer colleagues will leverage FARSIGHT and contribute code. At the same time, we hope that our life sciences colleagues will open our eyes to new problems and grand opportunities. Our goal is to foster a cross-disciplinary sharing of knowledge across the communities. We're all in this together. | ||
| + | |||
| + | '''Open Data & Imaging Protocols: ''' When completed, FARSIGHT will disseminate several high-quality datasets to the community to foster innovation and collaboration. We hope that our developer colleagues will use these datasets to advance the state of the art, and contribute their code to FARSIGHT. At the same time, we hope that our life sciences colleagues will post datastes to open the developers' eyes to new problems and applications. Click on the "OMERO Database" Link on the left sidebar to reach our image database. We are also sharing the specimen preparation and imaging protocols that are used to generate the images. Click on the "Imaging Protocols" tab on the left to learn more. | ||
| + | |||
| + | '''An Emphasis on Associative Image Analysis: ''' The FARSIGHT project is developing automated computational tools that can extract meaningful measurements from the complex and voluminous data generated by modern optical microscopes. Automation is important, but not our sole motivating force. We are interested in advancing a '''systems oriented''' understanding of complex and dynamic tissue microenvironments. This calls for a particular emphasis on quantifying, representing, and analyzing spatial and temporal associations among structural and functional tissue entities. This is the growing field of '''Biological Image Informatics'''[[http://en.wikipedia.org/wiki/Bioimage_informatics]]. | ||
| + | |||
| + | '''An Emphasis on Efficient Validation: ''' Ensuring the validity of the results on an operational basis is of utmost importance to this project. A closely related topic is performance assessment/profiling of automated algorithms. These activities are essential to enable much more widespread adoption of automated image analysis methods in biological investigations. Traditional validation methodologies are expensive to implement and do not provide sufficient performance data. To address this limitation, we are advancing the state of the art in validation methodologies. Our streamlined Pattern Analysis Aided Cluster Edit Based validation (PACE) methodology enables users to validate segmentation and classification results 'on the fly' with minimal effort. Visit the [[Validation Methods ]] page for more information. | ||
| + | |||
| + | '''Parallel Computation: ''' When completed, the FARSIGHT toolkit will enable us to take advantage of multi-core, multi-processor, and cluster computers. All routines are written in basic languages like C++ and Python so they can be run on most computers. Key time-consuming routines are being written using open parallel computing standards like MPI. For algorithms that are amenable to stream processing, we take advantage of graphics processors (GPUs) for desktop parallel processing. Some of our automated vessel segmentation routines are available in GPU accelerated versions. To learn more about GPU's click here [http://www.gpgpu.org GPGPU.ORG]. For some aspects of this project, we use one of the world's largest supercomputers that happens to be on the RPI campus. For more information, click here [http://www.rpi.edu/research/ccni/ CCNI]. | ||
| + | |||
| + | '''An emphasis on fluorescence microscopy: ''' The letter "F" in FARSIGHT stands for fluorescence, a molecular imaging technique that has revolutionized microscopy. Importantly, it allows us to greatly simplify the task of biological image analysis. | ||
| + | |||
| + | == RECENTLY ADDED PAGES == | ||
| + | * [[NucleusEditor|Nucleus Editor]] | ||
| + | * [[Seed_Editor|Seed Editor]] | ||
| + | * [[Segmentation Viewer]] | ||
| + | * [[Features|Intrinsic and Associative Features]] | ||
| + | * [[Registration page|3-D Registration & Montage Synthesis]] | ||
| + | * [[TissueNets Program|TissueNets Graph Builder for Secondary Associations]] | ||
| + | * [[Trace Editor| Program for inspecting and Editing Neurite/Vessel Traces in 3D]] | ||
| + | * [[MDL Neuron Modeling|Automated Complexity-constrained Segmentation of Neurites and Dendritic Spines]] | ||
| + | * [[3D Subcellular Location Features]] | ||
| + | * [[Vessel Laminae Segmentation|Automated Segmentation of Vasculature When the Vessel Laminae are Labeled & Imaged ]] | ||
| + | * [[Nuclear Segmentation|Program for Automated 2D/3D Segmentation of Cell Nuclei]] | ||
| + | * [[Algorithmic Information Theoretic Prediction & Discovery]] | ||
| + | * [[Object level association]] | ||
| + | * [[Histopathology|Histopathology: Quantifying Biomarkers at the Nucleus/Cellular Scale]] | ||
| + | * [[Tracking|5-D Image Analysis]] | ||
| + | * [[SuperEllipseTrace3D| Robust 3D Vasculature Tracing using Superellipsoids ]] | ||
| + | * [[Common_Tracing_Output_Format|Common Tracing Output Format]] | ||
| + | * [[Dendritic_Spine_Segmentation|Dendritic Spine Segmentation]] | ||
| + | * [[ITK_Pre-Processing_Algorithm_Wrappers_in_Python|ITK Pre-Processing Algorithm Wrappers in Python]] | ||
| + | * [[Worm Analysis System|Worm Analysis System: Automated Tracking and Analysis of C. ''elegans'']] | ||
| + | * [[Open Snake Tracing System|The Tracing System: A Broadly Applicable Neuron/Vessel Reconstruction System with Parallel Implementations]] | ||
| + | * [[Harmonic Co-clustering Heatmap|Harmonic Co-clustering Heatmap]] | ||
| + | * [[Automated Profiling of Individual Cell-Cell Interactions from High-throughput Time-lapse Imaging Microscopy in Nanowell Grids (TIMING)]] | ||
| + | * [[Morphologically Constrained Spectral Unmixing by Dictionary Learning for Multiplex Fluorescence Microscopy]] | ||
| + | |||
| + | == RECENT PUBLICATIONS == | ||
| + | |||
| + | [1] Bjornsson CS, Gang Lin, Al-Kofahi Y, Narayanaswamy A, Smith KL, Shain W, Roysam B. Associative image analysis: a method for automated quantification of 3D multi-parameter images of brain tissue. J. Neurosci. Methods, 170(1):165-78, 2008. | ||
| + | |||
| + | [2] Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell. Sep 11;3(3):289-300, 2008. | ||
| + | |||
| + | [3] Ying Chen, Ena Ladi, Paul Herzmark, Ellen Robey, and Badrinath Roysam, “Automated 5-D Analysis of Cell Migration and Interaction in the Thymic Cortex from Time-Lapse Sequences of 3-D Multi-channel Multi-photon Images” , Journal of Immunological Methods, 340(1):65-80, 2009. | ||
| + | |||
| + | [4] Ena Ladi, Tanja Schwickert, Tatyana Chtanova, Ying Chen, Paul Herzmark, Xinye Yin, Holly Aaron, Shiao Wei Chan, Martin Lipp, Badrinath Roysam and Ellen A. Robey, “Thymocyte-dendritic cell interactions near sources of CCR7 ligands in the thymic cortex,” Journal of Immunology 181(10):7014-23, 2008. | ||
| + | |||
| + | [5] Andrew R. Cohen, Christopher Bjornsson, Sally Temple, Gary Banker, and Badrinath Roysam, “Automatic Summarization of Changes in Biological Image Sequences using Algorithmic Information Theory” (in press, ePub available) IEEE Transactions on Pattern Analysis and Machine Intelligence, 2008. | ||
| + | |||
| + | [6] Padfield D, Rittscher J, Thomas N, Roysam B. Spatio-temporal cell cycle phase analysis using level sets and fast marching methods. Med Image Analysis, vol. 13, issue 1, pp. 143-155, February 2009. | ||
| + | |||
| + | [7] Arunachalam Narayanaswamy, Saritha Dwarakapuram, Christopher S. Bjornsson, Barbara M. Cutler, William Shain, Badrinath Roysam, Robust Adaptive 3-D Segmentation of Vessel Laminae from Fluorescence Confocal Microscope Images & Parallel GPU Implementation, (accepted, in press), IEEE Transactions on Medical Imaging, March 2009. | ||
Latest revision as of 19:08, 22 June 2016
Welcome to the FARSIGHT Project Wiki
This Project is Developing Quantitative Tools for Studying Complex and Dynamic Biological Microenvironments from 4D/5D Microscopy Data
The goal of the FARSIGHT project is to develop and disseminate a next-generation toolkit of image analysis methods to enable quantitative studies of complex & dynamic tissue microenvironments that are imaged by modern optical microscopes. Examples of such microenvironments include brain tissue, stem cell niches, developing embryonic tissue, immune system components, and tumors. Progress in mapping these microenvironments is much too slow compared to the need. Our knowledge of these systems has been painstakingly “pieced together” from large numbers of fixed, 2-D images of specimens revealing a small fraction of the molecular 'players' involved. The goal of this project is to help accelerate progress by: (i) harnessing the power of modern microscopy to help see the microenvironments in a much more detailed, direct, and comprehensive manner; and (ii) computational tools to analyze the multi-dimensional data produced by these microscopes.
Powered by this new Golden Age of optical microscopy: Modern optical microscopes can capture multi-dimensional images of tissue microenvironments. First of all, these microscopes can record three-dimensional (x,y,z) images of thick, intact slices that are more realistic compared to thin slices. Next, they can record multiple structures simultaneously in a manner that preserves their spatial inter-relationships. This allows us to make associative measurements in addition to traditional morphological measurements (we call them intrinsic measurements). Such four-dimensional imaging (x,y,z,λ) is usually accomplished using multiple fluorescent labels that tag the structures of interest with a high degree of molecular specificity. Finally, it is now possible to capture such 3-D multi-channel images of living systems in the form of a time-lapse movie (image sequence (x,y,z,t)) that reveals dynamic processes in the tissues. Using all of the available imaging dimensions (x,y,z,λ,t), we can now observe living processes in their native tissue habitat. Ongoing progress in this field is producing microscopes that can resolve much finer structures, produce images much faster, and on a much larger scale. In the future, one can expect further growth in the number of possible dimensions. For instance, fluorescence lifetimes indicate molecular nano-environments, and the inclusion of additional modalities such as phase, polarization and non-linear scatter will undoubtedly provide additional data. Click here to learn more about the rationale for multi-dimensional microscopy. To learn more about optical microscopy, [Click Here]
It's a Toolkit, not a Software package: We draw a distinction between these two words. A software package is a self-contained and tightly integrated software system that provides a defined set of services. A toolkit, on the other hand, is a collection of software modules with a set of standardized interfaces. To solve a given image analysis task, you can choose the right set of modules, and stitch them together using a scripting language ([Python] in our case). Toolkits are easier to build and maintain (especially for academic laboratories like us), and more versatile since we cannot foresee all possible applications that FARSIGHT will encounter in the future. Click here to learn more about the FARSIGHT Toolkit.
The FARSIGHT Framework: The FARSIGHT Framework serves as a conceptual guide to users and developers alike. The toolkit can be used to implement the framework by scripting modules together using the Python language. Click here to learn more about our long-term goals.
Open Source Software: FARSIGHT is an open source toolkit, and you can view our works in progress. You can also contribute to this project. FARSIGHT draws upon major open source toolkits especially the [Insight Toolkit (ITK)], the [Visualization Toolkit (VTK)], [Open Microscopy Environment (OME]), and various others. The FARSIGHT community is composed of users in the life sciences and developers in the computational sciences (happily, many colleagues seem at ease in both categories!). We hope that our developer colleagues will leverage FARSIGHT and contribute code. At the same time, we hope that our life sciences colleagues will open our eyes to new problems and grand opportunities. Our goal is to foster a cross-disciplinary sharing of knowledge across the communities. We're all in this together.
Open Data & Imaging Protocols: When completed, FARSIGHT will disseminate several high-quality datasets to the community to foster innovation and collaboration. We hope that our developer colleagues will use these datasets to advance the state of the art, and contribute their code to FARSIGHT. At the same time, we hope that our life sciences colleagues will post datastes to open the developers' eyes to new problems and applications. Click on the "OMERO Database" Link on the left sidebar to reach our image database. We are also sharing the specimen preparation and imaging protocols that are used to generate the images. Click on the "Imaging Protocols" tab on the left to learn more.
An Emphasis on Associative Image Analysis: The FARSIGHT project is developing automated computational tools that can extract meaningful measurements from the complex and voluminous data generated by modern optical microscopes. Automation is important, but not our sole motivating force. We are interested in advancing a systems oriented understanding of complex and dynamic tissue microenvironments. This calls for a particular emphasis on quantifying, representing, and analyzing spatial and temporal associations among structural and functional tissue entities. This is the growing field of Biological Image Informatics[[1]].
An Emphasis on Efficient Validation: Ensuring the validity of the results on an operational basis is of utmost importance to this project. A closely related topic is performance assessment/profiling of automated algorithms. These activities are essential to enable much more widespread adoption of automated image analysis methods in biological investigations. Traditional validation methodologies are expensive to implement and do not provide sufficient performance data. To address this limitation, we are advancing the state of the art in validation methodologies. Our streamlined Pattern Analysis Aided Cluster Edit Based validation (PACE) methodology enables users to validate segmentation and classification results 'on the fly' with minimal effort. Visit the Validation Methods page for more information.
Parallel Computation: When completed, the FARSIGHT toolkit will enable us to take advantage of multi-core, multi-processor, and cluster computers. All routines are written in basic languages like C++ and Python so they can be run on most computers. Key time-consuming routines are being written using open parallel computing standards like MPI. For algorithms that are amenable to stream processing, we take advantage of graphics processors (GPUs) for desktop parallel processing. Some of our automated vessel segmentation routines are available in GPU accelerated versions. To learn more about GPU's click here GPGPU.ORG. For some aspects of this project, we use one of the world's largest supercomputers that happens to be on the RPI campus. For more information, click here CCNI.
An emphasis on fluorescence microscopy: The letter "F" in FARSIGHT stands for fluorescence, a molecular imaging technique that has revolutionized microscopy. Importantly, it allows us to greatly simplify the task of biological image analysis.
RECENTLY ADDED PAGES
- Nucleus Editor
- Seed Editor
- Segmentation Viewer
- Intrinsic and Associative Features
- 3-D Registration & Montage Synthesis
- TissueNets Graph Builder for Secondary Associations
- Program for inspecting and Editing Neurite/Vessel Traces in 3D
- Automated Complexity-constrained Segmentation of Neurites and Dendritic Spines
- 3D Subcellular Location Features
- Automated Segmentation of Vasculature When the Vessel Laminae are Labeled & Imaged
- Program for Automated 2D/3D Segmentation of Cell Nuclei
- Algorithmic Information Theoretic Prediction & Discovery
- Object level association
- Histopathology: Quantifying Biomarkers at the Nucleus/Cellular Scale
- 5-D Image Analysis
- Robust 3D Vasculature Tracing using Superellipsoids
- Common Tracing Output Format
- Dendritic Spine Segmentation
- ITK Pre-Processing Algorithm Wrappers in Python
- Worm Analysis System: Automated Tracking and Analysis of C. elegans
- The Tracing System: A Broadly Applicable Neuron/Vessel Reconstruction System with Parallel Implementations
- Harmonic Co-clustering Heatmap
- Automated Profiling of Individual Cell-Cell Interactions from High-throughput Time-lapse Imaging Microscopy in Nanowell Grids (TIMING)
- Morphologically Constrained Spectral Unmixing by Dictionary Learning for Multiplex Fluorescence Microscopy
RECENT PUBLICATIONS
[1] Bjornsson CS, Gang Lin, Al-Kofahi Y, Narayanaswamy A, Smith KL, Shain W, Roysam B. Associative image analysis: a method for automated quantification of 3D multi-parameter images of brain tissue. J. Neurosci. Methods, 170(1):165-78, 2008.
[2] Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell Stem Cell. Sep 11;3(3):289-300, 2008.
[3] Ying Chen, Ena Ladi, Paul Herzmark, Ellen Robey, and Badrinath Roysam, “Automated 5-D Analysis of Cell Migration and Interaction in the Thymic Cortex from Time-Lapse Sequences of 3-D Multi-channel Multi-photon Images” , Journal of Immunological Methods, 340(1):65-80, 2009.
[4] Ena Ladi, Tanja Schwickert, Tatyana Chtanova, Ying Chen, Paul Herzmark, Xinye Yin, Holly Aaron, Shiao Wei Chan, Martin Lipp, Badrinath Roysam and Ellen A. Robey, “Thymocyte-dendritic cell interactions near sources of CCR7 ligands in the thymic cortex,” Journal of Immunology 181(10):7014-23, 2008.
[5] Andrew R. Cohen, Christopher Bjornsson, Sally Temple, Gary Banker, and Badrinath Roysam, “Automatic Summarization of Changes in Biological Image Sequences using Algorithmic Information Theory” (in press, ePub available) IEEE Transactions on Pattern Analysis and Machine Intelligence, 2008.
[6] Padfield D, Rittscher J, Thomas N, Roysam B. Spatio-temporal cell cycle phase analysis using level sets and fast marching methods. Med Image Analysis, vol. 13, issue 1, pp. 143-155, February 2009.
[7] Arunachalam Narayanaswamy, Saritha Dwarakapuram, Christopher S. Bjornsson, Barbara M. Cutler, William Shain, Badrinath Roysam, Robust Adaptive 3-D Segmentation of Vessel Laminae from Fluorescence Confocal Microscope Images & Parallel GPU Implementation, (accepted, in press), IEEE Transactions on Medical Imaging, March 2009.